6XFD
 
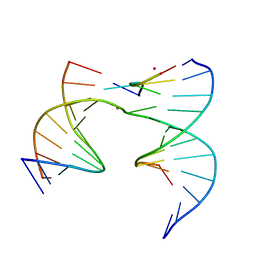 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J20 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*GP*TP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*CP*CP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-06-15 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.101 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6LF5
 
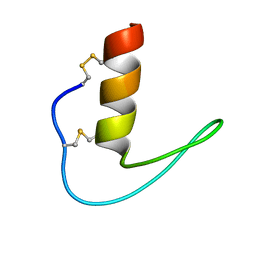 | | The solution structure of ShSPI | | Descriptor: | ShSPI | | Authors: | Luan, N, Rong, M.Q, Liu, J.X, Lai, R. | | Deposit date: | 2019-11-29 | | Release date: | 2020-12-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Identification and Characterization of ShSPI, a Kazal-Type Elastase Inhibitor from the Venom of Scolopendra Hainanum .
Toxins, 11, 2019
|
|
6XGJ
 
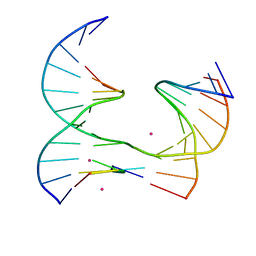 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J32 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*TP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*GP*AP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-06-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.071 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XQ9
 
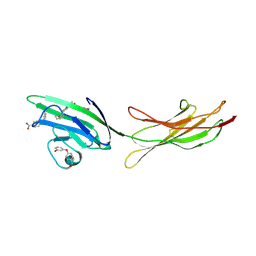 | | Receptor for Advanced Glycation End Products VC1 domain in complex with Fragments 1 & 13 | | Descriptor: | 4-(3-hydroxyphenoxy)benzoic acid, 7-methyl-3-phenyl-1H-indole-2-carboxylic acid, ACETATE ION, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
6XFG
 
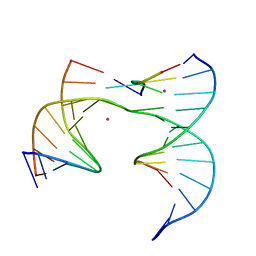 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J23 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*TP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*GP*GP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-06-15 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.053 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6KVG
 
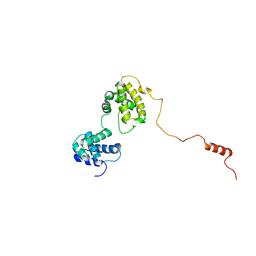 | | The solution structure of human Orc6 | | Descriptor: | Origin recognition complex subunit 6 | | Authors: | Liu, C, Xu, N, You, Y, Zhu, G. | | Deposit date: | 2019-09-04 | | Release date: | 2020-09-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA replication origin recognition by human Orc6 protein binding with DNA.
Nucleic Acids Res., 48, 2020
|
|
6XFW
 
 | | Self-assembly of a 3D DNA crystal lattice (4x5 junction version) containing the J24 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*CP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*GP*GP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-06-16 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.053 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
6XO6
 
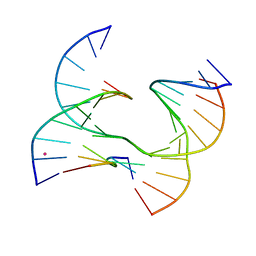 | | Self-assembly of a 3D DNA crystal lattice (4x6 junction version) containing the J28 immobile Holliday junction | | Descriptor: | CACODYLATE ION, DNA (5'-D(*GP*AP*GP*CP*AP*GP*AP*CP*AP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*GP*AP*GP*TP*GP*TP*GP*TP*CP*TP*GP*C)-3'), ... | | Authors: | Simmons, C.R, MacCulloch, T, Stephanopoulos, N, Yan, H. | | Deposit date: | 2020-07-06 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.108 Å) | | Cite: | The influence of Holliday junction sequence and dynamics on DNA crystal self-assembly.
Nat Commun, 13, 2022
|
|
1EJ4
 
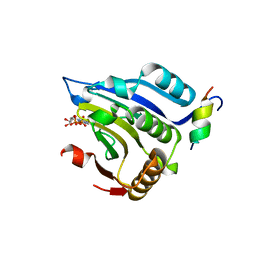 | | COCRYSTAL STRUCTURE OF EIF4E/4E-BP1 PEPTIDE | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, EUKARYOTIC INITIATION FACTOR 4E, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E BINDING PROTEIN 1 | | Authors: | Marcotrigiano, J, Gingras, A.-C, Sonenberg, N, Burley, S.K. | | Deposit date: | 2000-02-28 | | Release date: | 2000-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G.
Mol.Cell, 3, 1999
|
|
6XQ1
 
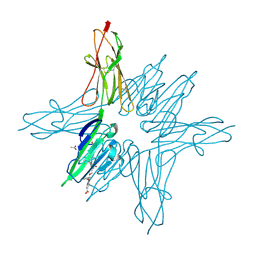 | | Receptor for Advanced Glycation End Products VC1 domain in complex with 3-(3-((4-(4-carboxyphenoxy)benzyl)oxy)phenyl)-1H-indole-2-carboxylic acid | | Descriptor: | 3-[3-({[3-(4-carboxyphenoxy)phenyl]methoxy}methyl)phenyl]-1H-indole-2-carboxylic acid, ACETATE ION, Advanced glycosylation end product-specific receptor, ... | | Authors: | Salay, L.E, Kozlyuk, N, Gilston, B.A, Gogliotti, R.D, Christov, P.P, Kim, K, Ovee, M, Waterson, A.G, Chazin, W.J. | | Deposit date: | 2020-07-09 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | A fragment-based approach to discovery of Receptor for Advanced Glycation End products inhibitors.
Proteins, 89, 2021
|
|
1EDW
 
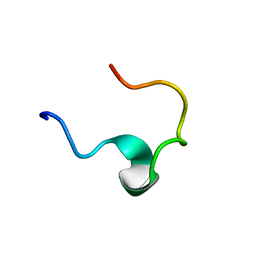 | | SOLUTION STRUCTURE OF THIRD INTRADISKAL LOOP OF BOVINE RHODOPSIN (RESIDUES 268-293) | | Descriptor: | RHODOPSIN | | Authors: | Yeagle, P.L, Salloum, A, Chopra, A, Bhawsar, N, Ali, L. | | Deposit date: | 2000-01-28 | | Release date: | 2000-08-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of the intradiskal loops and amino terminus of the G-protein receptor, rhodopsin.
J.Pept.Res., 55, 2000
|
|
1EKQ
 
 | |
1ET0
 
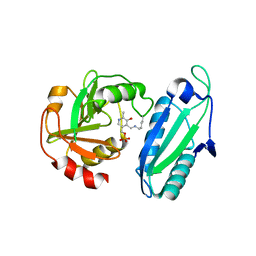 | | CRYSTAL STRUCTURE OF AMINODEOXYCHORISMATE LYASE FROM ESCHERICHIA COLI | | Descriptor: | 4-AMINO-4-DEOXYCHORISMATE LYASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Nakai, T, Mizutani, H, Miyahara, I, Hirotsu, K, Takeda, S, Jhee, K.H, Yoshimura, T, Esaki, N. | | Deposit date: | 2000-04-12 | | Release date: | 2000-07-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of 4-amino-4-deoxychorismate lyase from Escherichia coli.
J.Biochem.(Tokyo), 128, 2000
|
|
6LYY
 
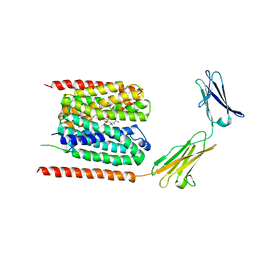 | | Cryo-EM structure of the human MCT1/Basigin-2 complex in the presence of anti-cancer drug candidate AZD3965 in the outward-open conformation. | | Descriptor: | 3-methyl-5-[[(4~{R})-4-methyl-4-oxidanyl-1,2-oxazolidin-2-yl]carbonyl]-6-[[5-methyl-3-(trifluoromethyl)-1~{H}-pyrazol-4-yl]methyl]-1-propan-2-yl-thieno[2,3-d]pyrimidine-2,4-dione, Basigin, Monocarboxylate transporter 1 | | Authors: | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | Deposit date: | 2020-02-16 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
6LS4
 
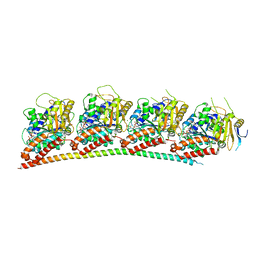 | | A novel anti-tumor agent S-40 in complex with tubulin | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-[(4-cyclopropylphenyl)sulfonylamino]-4-methyl-N-(pyridin-3-ylmethyl)benzamide, GLYCEROL, ... | | Authors: | Du, T, Lin, S, Ji, M, Xue, N, Liu, Y, Zhang, K, Lu, D, Chen, X, Xu, H. | | Deposit date: | 2020-01-17 | | Release date: | 2021-01-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel orally active microtubule destabilizing agent S-40 targets the colchicine-binding site and shows potent antitumor activity.
Cancer Lett., 495, 2020
|
|
1EDX
 
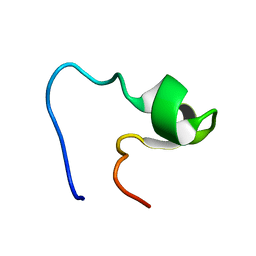 | | SOLUTION STRUCTURE OF AMINO TERMINUS OF BOVINE RHODOPSIN (RESIDUES 1-40) | | Descriptor: | RHODOPSIN | | Authors: | Yeagle, P.L, Salloum, A, Chopra, A, Bhawsar, N, Ali, L. | | Deposit date: | 2000-01-28 | | Release date: | 2000-08-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of the intradiskal loops and amino terminus of the G-protein receptor, rhodopsin.
J.Pept.Res., 55, 2000
|
|
6Y6K
 
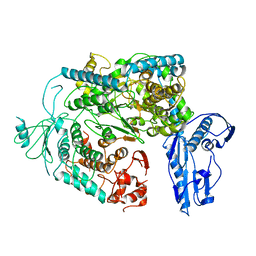 | | Cryo-EM structure of a Phenuiviridae L protein | | Descriptor: | MAGNESIUM ION, RNA-dependent RNA polymerase | | Authors: | Vogel, D, Thorkelsson, S.R, Quemin, E, Meier, K, Kouba, T, Gogrefe, N, Busch, C, Reindl, S, Guenther, S, Cusack, S, Gruenewald, K, Rosenthal, M. | | Deposit date: | 2020-02-26 | | Release date: | 2020-04-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Structural and functional characterization of the severe fever with thrombocytopenia syndrome virus L protein.
Nucleic Acids Res., 48, 2020
|
|
6Y16
 
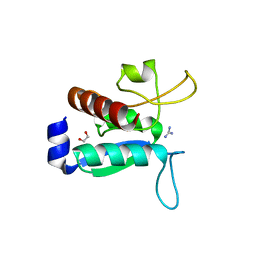 | | CRYSTAL STRUCTURE OF TMARGBP DOMAIN 1 IN COMPLEX WITH THE GUANIDINIUM ION | | Descriptor: | 1,2-ETHANEDIOL, Amino acid ABC transporter, periplasmic amino acid-binding protein,Amino acid ABC transporter, ... | | Authors: | Ruggiero, A, Balasco, N, Smaldone, G, Graziano, G, Vitagliano, L. | | Deposit date: | 2020-02-11 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Guanidinium binding to proteins: The intriguing effects on the D1 and D2 domains of Thermotoga maritima Arginine Binding Protein and a comprehensive analysis of the Protein Data Bank.
Int.J.Biol.Macromol., 163, 2020
|
|
6M5S
 
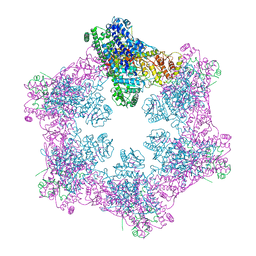 | | The coordinates of the apo hexameric terminase complex | | Descriptor: | Tripartite terminase subunit 1, Tripartite terminase subunit 2, Tripartite terminase subunit 3, ... | | Authors: | Yang, Y.X, Yang, P, Wang, N, Chen, Z.H, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | Deposit date: | 2020-03-11 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
6M8F
 
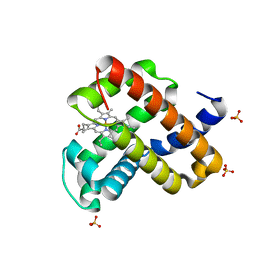 | | Engineered sperm whale myoglobin-based carbene transferase | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, ... | | Authors: | Bacik, J.P, Ando, N, Fasan, R. | | Deposit date: | 2018-08-21 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Origin of high stereocontrol in olefin cyclopropanation catalyzed by an engineered carbene transferase.
Acs Catalysis, 9, 2019
|
|
1EJ5
 
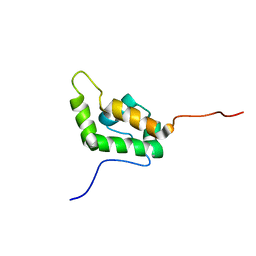 | | SOLUTION STRUCTURE OF THE AUTOINHIBITED CONFORMATION OF WASP | | Descriptor: | WISKOTT-ALDRICH SYNDROME PROTEIN | | Authors: | Kim, A.S, Kakalis, L.T, Abdul-Manan, N, Liu, G.A, Rosen, M.K. | | Deposit date: | 2000-02-29 | | Release date: | 2000-04-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Autoinhibition and activation mechanisms of the Wiskott-Aldrich syndrome protein.
Nature, 404, 2000
|
|
6M49
 
 | | cryo-EM structure of Scap/Insig complex in the present of 25-hydroxyl cholesterol. | | Descriptor: | 25-HYDROXYCHOLESTEROL, Insulin-induced gene 2 protein, Sterol regulatory element-binding protein cleavage-activating protein,Sterol regulatory element-binding protein cleavage-activating protein | | Authors: | Yan, R, Cao, P, Song, W, Qian, H, Du, X, Coates, H.W, Zhao, X, Li, Y, Gao, S, Gong, X, Liu, X, Sui, J, Lei, J, Yang, H, Brown, A.J, Zhou, Q, Yan, C, Yan, N. | | Deposit date: | 2020-03-06 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A structure of human Scap bound to Insig-2 suggests how their interaction is regulated by sterols.
Science, 371, 2021
|
|
6LS8
 
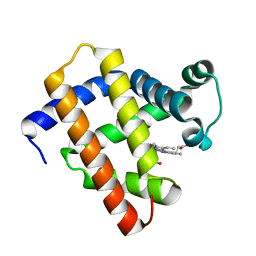 | | The monomeric structure of G80A/H81A/H82A myoglobin | | Descriptor: | Myoglobin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nagao, S, Suda, A, Kobayashi, H, Shibata, N, Higuchi, Y, Hirota, S. | | Deposit date: | 2020-01-17 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Thermodynamic Control of Domain Swapping by Modulating the Helical Propensity in the Hinge Region of Myoglobin.
Chem Asian J, 15, 2020
|
|
6M5U
 
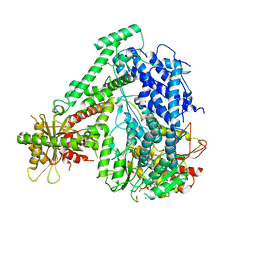 | | The coordinates of the monomeric terminase complex in the presence of the ADP-BeF3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Yang, Y.X, Yang, P, Wang, N, Zhu, L, Zhou, Z.H, Rao, Z.H, Wang, X.X. | | Deposit date: | 2020-03-11 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Architecture of the herpesvirus genome-packaging complex and implications for DNA translocation.
Protein Cell, 11, 2020
|
|
1EDV
 
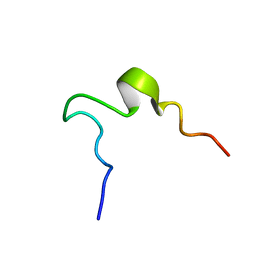 | | SOLUTION STRUCTURE OF 2ND INTRADISKAL LOOP OF BOVINE RHODOPSIN (RESIDUES 172-205) | | Descriptor: | RHODOPSIN | | Authors: | Yeagle, P.L, Salloum, A, Chopra, A, Bhawsar, N, Ali, L. | | Deposit date: | 2000-01-28 | | Release date: | 2000-08-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structures of the intradiskal loops and amino terminus of the G-protein receptor, rhodopsin.
J.Pept.Res., 55, 2000
|
|
