7PBQ
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s0+A [t2 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Goessweiner-Mohr, N, Fahrenkamp, D, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
1BGK
 
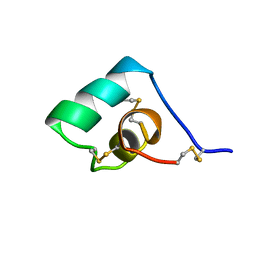 | | SEA ANEMONE TOXIN (BGK) WITH HIGH AFFINITY FOR VOLTAGE DEPENDENT POTASSIUM CHANNEL, NMR, 15 STRUCTURES | | Descriptor: | BGK | | Authors: | Dauplais, M, Lecoq, A, Song, J, Cotton, J, Jamin, N, Gilquin, B, Roumestand, C, Vita, C, Harvey, A, Menez, A. | | Deposit date: | 1996-05-08 | | Release date: | 1997-01-27 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | On the convergent evolution of animal toxins. Conservation of a diad of functional residues in potassium channel-blocking toxins with unrelated structures.
J.Biol.Chem., 272, 1997
|
|
7PBS
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s0+A [t1 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvA, Holliday junction ATP-dependent DNA helicase RuvB, ... | | Authors: | Goessweiner-Mohr, N, Fahrenkamp, D, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
8ERE
 
 | |
8ERG
 
 | |
7PBR
 
 | | RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s0-A [t2 dataset] | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvB, MAGNESIUM ION, ... | | Authors: | Goessweiner-Mohr, N, Fahrenkamp, D, Wald, J, Marlovits, T.C. | | Deposit date: | 2021-08-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mechanism of AAA+ ATPase-mediated RuvAB-Holliday junction branch migration.
Nature, 609, 2022
|
|
8ERI
 
 | |
8ERF
 
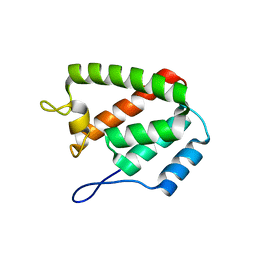 | |
1R4G
 
 | | Solution structure of the Sendai virus protein X C-subdomain | | Descriptor: | RNA polymerase alpha subunit | | Authors: | Blanchard, L, Tarbouriech, N, Blackledge, M, Timmins, P, Burmeister, W.P, Ruigrok, R.W, Marion, D. | | Deposit date: | 2003-10-06 | | Release date: | 2004-03-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of the nucleocapsid-binding domain of the Sendai virus phosphoprotein in solution
Virology, 319, 2004
|
|
7Q10
 
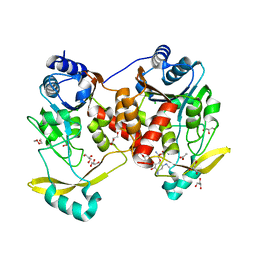 | | Acetyltrasferase(3) type IIIa in complex with 3-N-methyl-nemycin B | | Descriptor: | 3N methyl nemycin B, ACETATE ION, Aminoglycoside N(3)-acetyltransferase III, ... | | Authors: | Pontillo, N, Guskov, A. | | Deposit date: | 2021-10-17 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | 3-N-alkylation in aminoglycoside antibiotic neomycin B overcomes bacterial resistance mediated by acetyltransferase (3) IIIa
To Be Published
|
|
7PRL
 
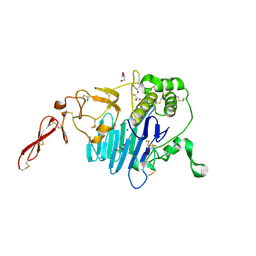 | | MUC2 D1 with Cu(II) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, GLYCEROL, ... | | Authors: | Reznik, N, Fass, D. | | Deposit date: | 2021-09-22 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Intestinal mucin is a chaperone of multivalent copper.
Cell, 185, 2022
|
|
6W5S
 
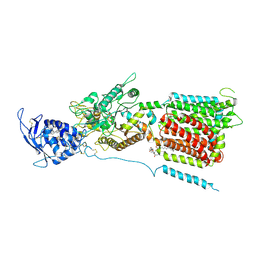 | | NPC1 structure in GDN micelles at pH 8.0 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Yan, N, Qian, H.W, Wu, X.L. | | Deposit date: | 2020-03-13 | | Release date: | 2020-06-17 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural Basis of Low-pH-Dependent Lysosomal Cholesterol Egress by NPC1 and NPC2.
Cell, 182, 2020
|
|
6VTU
 
 | | DH717.1 Fab monomer in complex with man9 glycan | | Descriptor: | DH717.1 heavy chain, DH717.1 light chain, GLYCEROL, ... | | Authors: | Fera, D, Bronkema, N. | | Deposit date: | 2020-02-13 | | Release date: | 2020-12-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies.
Cell, 184, 2021
|
|
6VX7
 
 | | bestrophin-2 Ca2+-bound state (5 mM Ca2+) | | Descriptor: | Bestrophin, CALCIUM ION, CHLORIDE ION | | Authors: | Owji, A.P, Zhao, Q, Ji, C, Kittredge, A, Hopiavuori, A, Fu, Z, Ward, N, Clarke, O, Shen, Y, Zhang, Y, Hendrickson, W.A, Yang, T. | | Deposit date: | 2020-02-21 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.36 Å) | | Cite: | Structural and functional characterization of the bestrophin-2 anion channel.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6VYP
 
 | | Crystal structure of the LSD1/CoREST histone demethylase bound to its nucleosome substrate | | Descriptor: | DNA (191-MER), FLAVIN-ADENINE DINUCLEOTIDE, Histone H2A type 1, ... | | Authors: | Kim, S, Zhu, J, Eek, P, Yennawar, N, Song, T. | | Deposit date: | 2020-02-27 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.99 Å) | | Cite: | Crystal Structure of the LSD1/CoREST Histone Demethylase Bound to Its Nucleosome Substrate.
Mol.Cell, 78, 2020
|
|
7PP1
 
 | |
7Q1D
 
 | | Acetyltrasferase(3) type IIIa in complex with 3-N-methyl-nemycin B | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Aminoglycoside N(3)-acetyltransferase III, CHLORIDE ION, ... | | Authors: | Pontillo, N, Guskov, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | 3-N-alkylation in aminoglycoside antibiotic neomycin B overcomes bacterial resistance mediated by acetyltransferase (3) IIIa
To Be Published
|
|
8ED5
 
 | |
6VP0
 
 | | Human Diacylglycerol Acyltransferase 1 in complex with oleoyl-CoA | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Diacylglycerol O-acyltransferase 1, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Wang, L, Qian, H, Han, Y, Nian, Y, Ren, Z, Zhang, H, Hu, L, Prasad, B.V.V, Yan, N, Zhou, M. | | Deposit date: | 2020-02-01 | | Release date: | 2020-05-13 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and mechanism of human diacylglycerol O-acyltransferase 1.
Nature, 581, 2020
|
|
7Q0Q
 
 | | Acetyltrasferase(3) type IIIa in complex with 3-N-methyl-nemycin B | | Descriptor: | 3N methyl nemycin B, ACETATE ION, Aminoglycoside N(3)-acetyltransferase III, ... | | Authors: | Pontillo, N, Guskov, A. | | Deposit date: | 2021-10-16 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | The 3-N-alkylation of the neomycin B outmaneuvers the aminoglycoside resistant enzyme acetyltransferase(3)IIIa via an unexpected mechanism
To Be Published
|
|
8F5X
 
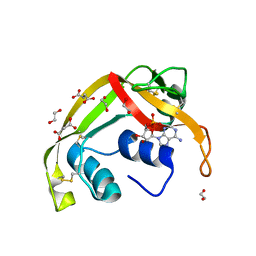 | | Crystal structure of human eosinophil-derived neurotoxin (EDN, ribonuclease 2) in complex with 5'-adenosine monophosphate (AMP) | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Non-secretory ribonuclease, ... | | Authors: | Tran, T.T.Q, Pham, N.T.H, Calmettes, C, Doucet, N. | | Deposit date: | 2022-11-15 | | Release date: | 2023-11-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ancestral sequence reconstruction dissects structural and functional differences among eosinophil ribonucleases.
J.Biol.Chem., 300, 2024
|
|
6VO3
 
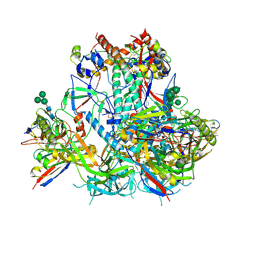 | | AMC009 SOSIP.v4.2 in complex with PGV04 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AMC009 SOSIP.v4.2 envelope glycoprotein gp120, ... | | Authors: | Cottrell, C.A, de Val, N, Ward, A.B. | | Deposit date: | 2020-01-29 | | Release date: | 2020-09-23 | | Last modified: | 2020-12-09 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Neutralizing Antibody Responses Induced by HIV-1 Envelope Glycoprotein SOSIP Trimers Derived from Elite Neutralizers.
J.Virol., 94, 2020
|
|
8ESU
 
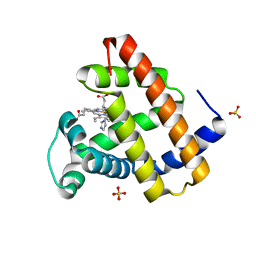 | | Myoglobin variant Mb-imi complex | | Descriptor: | IMIDAZOLE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Bacik, J.P, Fasan, R, Ando, N. | | Deposit date: | 2022-10-14 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Mechanistic manifold in a hemoprotein-catalyzed cyclopropanation reaction with diazoketone.
Nat Commun, 14, 2023
|
|
7Q1X
 
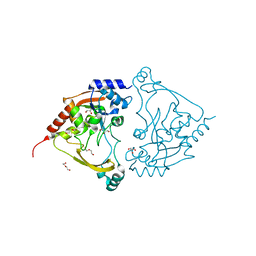 | | Acetyltrasferase(3) type IIIa in complex with neomycin B | | Descriptor: | Aminoglycoside N(3)-acetyltransferase III, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pontillo, N, Guskov, A. | | Deposit date: | 2021-10-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The 3-N-alkylation of the neomycin B outmaneuvers the aminoglycoside resistant enzyme acetyltransferase(3)IIIa via an unexpected mechanism
To Be Published
|
|
6VRB
 
 | | Cryo-EM structure of AcrVIA1-Cas13(crRNA) complex | | Descriptor: | AcrVIA1, CRISPR-associated endoribonuclease Cas13a, RNA (52-MER) | | Authors: | Jia, N, Meeske, A.J, Marraffini, L.A, Patel, D.J. | | Deposit date: | 2020-02-07 | | Release date: | 2020-06-10 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A phage-encoded anti-CRISPR enables complete evasion of type VI-A CRISPR-Cas immunity.
Science, 369, 2020
|
|
