7AA9
 
 | | Structure of SCOC pT13/pT15 LIR motif bound to GABARAPL1 | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 1, pT13/PT15 SCOC LIR | | Authors: | Lee, R, Mouilleron, S, Wirth, M, Zhang, W, O Reilly, N, Dhira, J, Tooze, S. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Phosphorylation of the LIR Domain of SCOC Modulates ATG8 Binding Affinity and Specificity.
J.Mol.Biol., 433, 2021
|
|
6MEN
 
 | | Crystal structure of a Tylonycteris bat coronavirus HKU4 macrodomain in complex with adenosine diphosphate glucose (ADP-glucose) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE-GLUCOSE, Replicase polyprotein 1ab | | Authors: | Hammond, R.G, Schormann, N, McPherson, R.L, Leung, A.K.L, Deivanayagam, C.C.S, Johnson, M.A. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | ADP-Ribose and Analogues bound to the DeMARylating Macrodomain from the Bat Coronavirus HKU4
Proc.Natl.Acad.Sci.USA, 2021
|
|
7AA8
 
 | | Structure of SCOC LIR bound to GABARAP | | Descriptor: | Chimera made of SCOC (6-23) + linker (GS) + GABARAP,Gamma-aminobutyric acid receptor-associated protein | | Authors: | Lee, R, Mouilleron, S, Wirth, M, Zhang, W, O Reilly, N, Dhira, J, Tooze, S. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Phosphorylation of the LIR Domain of SCOC Modulates ATG8 Binding Affinity and Specificity.
J.Mol.Biol., 433, 2021
|
|
7AA7
 
 | | Structure of SCOC pS12/pS18 LIR motif bound to GABARAPL1 | | Descriptor: | Gamma-aminobutyric acid receptor-associated protein-like 1, pS12/pS18 SCOC LIR, sulfoacetic acid | | Authors: | Lee, R, Mouilleron, S, Wirth, M, Zhang, W, O Reilly, N, Dhira, J, Tooze, S. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Phosphorylation of the LIR Domain of SCOC Modulates ATG8 Binding Affinity and Specificity.
J.Mol.Biol., 433, 2021
|
|
6LYM
 
 | | Crystal structure of D657A mutant of formylglycinamidine synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Sharma, N, Tanwar, A.S, Anand, R. | | Deposit date: | 2020-02-14 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Mechanism of Coordinated Gating and Signal Transduction in Purine Biosynthetic Enzyme Formylglycinamidine Synthetase.
Acs Catalysis, 12, 2022
|
|
6LYO
 
 | | Crystal Structure of H296A mutant of Formylglycinamidine Synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Sharma, N, Tanwar, A.S, Anand, R. | | Deposit date: | 2020-02-15 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Mechanism of Coordinated Gating and Signal Transduction in Purine Biosynthetic Enzyme Formylglycinamidine Synthetase.
Acs Catalysis, 12, 2022
|
|
6LYK
 
 | | Crystal Structure of R1263A mutant of Formylglycinamidine Synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLUTAMINE, ... | | Authors: | Sharma, N, Tanwar, A.S, Anand, R. | | Deposit date: | 2020-02-14 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of Coordinated Gating and Signal Transduction in Purine Biosynthetic Enzyme Formylglycinamidine Synthetase.
Acs Catalysis, 12, 2022
|
|
6LYL
 
 | | Crystal structure of S1052D mutant of Formylglycinamidine synthetase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Sharma, N, Tanwar, A.S, Anand, R. | | Deposit date: | 2020-02-14 | | Release date: | 2021-02-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of Coordinated Gating and Signal Transduction in Purine Biosynthetic Enzyme Formylglycinamidine Synthetase.
Acs Catalysis, 12, 2022
|
|
6LXV
 
 | | Cryo-EM structure of phosphoketolase from Bifidobacterium longum | | Descriptor: | CALCIUM ION, Phosphoketolase, THIAMINE DIPHOSPHATE | | Authors: | Nakata, K, Miyazaki, N, Yamaguchi, H, Hirose, M, Miyano, H, Mizukoshi, T, Kashiwagi, T, Iwasaki, K. | | Deposit date: | 2020-02-12 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | High-resolution structure of phosphoketolase from Bifidobacterium longum determined by cryo-EM single-particle analysis.
J.Struct.Biol., 214, 2022
|
|
6M6I
 
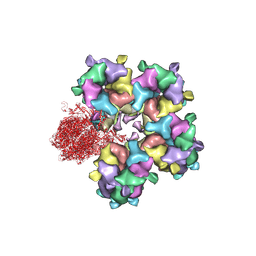 | | Structure of HSV2 B-capsid portal vertex | | Descriptor: | Coiled coils chain 1, Coiled coils chain 2, Major capsid protein, ... | | Authors: | Wang, X.X, Wang, N. | | Deposit date: | 2020-03-14 | | Release date: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Structures of the portal vertex reveal essential protein-protein interactions for Herpesvirus assembly and maturation.
Protein Cell, 11, 2020
|
|
6M7D
 
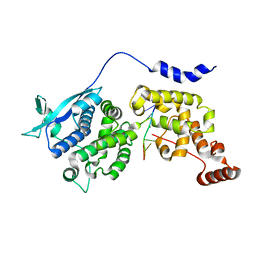 | | Structure of ncleoprotein of sendai virus | | Descriptor: | Nucleoprotein, RNA (5'-R(P*UP*UP*UP*UP*UP*U)-3') | | Authors: | Shen, Q, Shan, H, Zhang, N. | | Deposit date: | 2020-03-18 | | Release date: | 2021-03-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | structure of the nucleocapsid of sendai virus at 2.9 Angstroms resolution
To Be Published
|
|
6LQA
 
 | | voltage-gated sodium channel Nav1.5 with quinidine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Quinidine, Sodium channel protein type 5 subunit alpha | | Authors: | Yan, N, Li, Z, Pan, X, Huang, G. | | Deposit date: | 2020-01-13 | | Release date: | 2021-03-24 | | Last modified: | 2021-05-19 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Basis for Pore Blockade of the Human Cardiac Sodium Channel Na v 1.5 by the Antiarrhythmic Drug Quinidine*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6M6H
 
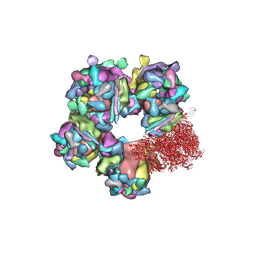 | | Structure of HSV2 C-capsid portal vertex | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Wang, X.X, Wang, N. | | Deposit date: | 2020-03-14 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures of the portal vertex reveal essential protein-protein interactions for Herpesvirus assembly and maturation.
Protein Cell, 11, 2020
|
|
6M6G
 
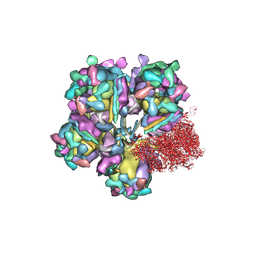 | | Structure of HSV2 viron capsid portal vertex | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Coiled coils, ... | | Authors: | Wang, X.X, Wang, N. | | Deposit date: | 2020-03-14 | | Release date: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (5.39 Å) | | Cite: | Structures of the portal vertex reveal essential protein-protein interactions for Herpesvirus assembly and maturation.
Protein Cell, 11, 2020
|
|
6M05
 
 | | Trimolecular G-quadruplex | | Descriptor: | DNA (5'-D(*GP*TP*TP*AP*GP*G)-3') | | Authors: | Jing, H.T, Fu, W.Q, Zhang, N. | | Deposit date: | 2020-02-20 | | Release date: | 2021-01-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structural study on the self-trimerization of d(GTTAGG) into a dynamic trimolecular G-quadruplex assembly preferentially in Na+ solution with a moderate K+ tolerance.
Nucleic Acids Res., 49, 2021
|
|
6WIB
 
 | | Next generation monomeric IgG4 Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin heavy constant gamma 4, ZINC ION | | Authors: | Oganesyan, V.Y, Shan, L, Dall'Acqua, W, van Dyk, N. | | Deposit date: | 2020-04-09 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | In vivo pharmacokinetic enhancement of monomeric Fc and monovalent bispecific designs through structural guidance.
Commun Biol, 4, 2021
|
|
6WMH
 
 | |
6WNA
 
 | | Next generation monomeric IgG4 Fc | | Descriptor: | Beta-2-microglobulin, IgG receptor FcRn large subunit p51, Immunoglobulin heavy constant gamma 4, ... | | Authors: | Oganesyan, V.Y, Shan, L, van Dyk, N, Dall'Acqua, W.F. | | Deposit date: | 2020-04-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | In vivo pharmacokinetic enhancement of monomeric Fc and monovalent bispecific designs through structural guidance.
Commun Biol, 4, 2021
|
|
6WOL
 
 | | Next generation monomeric IgG4 Fc bound to neonatal Fc receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, IgG receptor FcRn large subunit p51, ... | | Authors: | Oganesyan, V.Y, Shan, L, van Dyk, N, Dall'Acqua, W.F. | | Deposit date: | 2020-04-24 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | In vivo pharmacokinetic enhancement of monomeric Fc and monovalent bispecific designs through structural guidance.
Commun Biol, 4, 2021
|
|
6MGF
 
 | | untagged, wild-type LptB in complex with ADP | | Descriptor: | (3s,5s,7s)-N-{7-[(3-O-carbamoyl-6-deoxy-5-methyl-4-O-methyl-beta-D-gulopyranosyl)oxy]-4-hydroxy-8-methyl-2-oxo-2H-1-ben zopyran-3-yl}tricyclo[3.3.1.1~3,7~]decane-1-carboxamide, ADENOSINE-5'-DIPHOSPHATE, Lipopolysaccharide export system ATP-binding protein LptB, ... | | Authors: | Mandler, M.D, Owens, T.W, Ruiz, N, Kahne, D. | | Deposit date: | 2018-09-13 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.982 Å) | | Cite: | Bypassing the requirement for coupling of ATP binding and hydrolysis in the lipopolysaccharide ABC transporte
not published
|
|
6MJ3
 
 | | CRYSTAL STRUCTURE OF RHESUS MACAQUE (MACACA MULATTA) IGG1 Fc Fragment-Fc-GAMMA RECEPTOR III complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Igg1 Fc, ... | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2018-09-20 | | Release date: | 2019-08-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Decoding human-macaque interspecies differences in Fc-effector functions: The structural basis for CD16-dependent effector function in Rhesus macaques.
Front Immunol, 13, 2022
|
|
6MHM
 
 | | Crystal structure of human acid ceramidase in covalent complex with carmofur | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dementiev, A, Joachimiak, A, Doan, N. | | Deposit date: | 2018-09-18 | | Release date: | 2019-01-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.743 Å) | | Cite: | Molecular Mechanism of Inhibition of Acid Ceramidase by Carmofur.
J. Med. Chem., 62, 2019
|
|
6MGU
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus Anthracis in the complex with inhibitor Oxanosine monophosphate | | Descriptor: | 1,2-ETHANEDIOL, 5-[(Z)-(aminomethylidene)amino]-1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole-4-carboxylic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kim, Y, Maltseva, N, Yu, R, Hedstrom, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-14 | | Release date: | 2018-10-24 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus Anthracis in the complex with inhibitor Oxanosine monophosphate
To Be Published
|
|
6MGR
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor Oxanosine monophosphate | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-[(Z)-(aminomethylidene)amino]-1-(5-O-phosphono-beta-D-ribofuranosyl)-1H-imidazole-4-carboxylic acid, CHLORIDE ION, ... | | Authors: | Kim, Y, Maltseva, N, Yu, R, Hedstrom, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-09-14 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor Oxanosine Monophosphate
To Be Published
|
|
6MJC
 
 | |
