4YCP
 
 | | E. coli dihydrouridine synthase C (DusC) in complex with tRNATrp | | Descriptor: | FLAVIN MONONUCLEOTIDE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Byrne, R.T, Jenkins, H.T, Peters, D.T, Whelan, F, Stowell, J, Aziz, N, Kasatsky, P, Rodnina, M.V, Koonin, E.V, Konevega, A.L, Antson, A.A. | | Deposit date: | 2015-02-20 | | Release date: | 2015-04-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Major reorientation of tRNA substrates defines specificity of dihydrouridine synthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8IYR
 
 | |
8J59
 
 | |
8IYP
 
 | | Crystal structure of serine palmitoyltransferase soaked in 190 mM D-serine solution | | Descriptor: | 1,2-ETHANEDIOL, Serine palmitoyltransferase, [3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-SERINE | | Authors: | Takahashi, A, Murakami, T, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2023-04-05 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Crystal structure of Serine Palmitoyltransferase from Sphingobacterium multivorum
To Be Published
|
|
6FDD
 
 | | Crystal Structure of the HHD2 Domain of Whirlin | | Descriptor: | SULFATE ION, Whirlin | | Authors: | Delhommel, F, Cordier, F, Saul, F, Haouz, A, Wolff, N. | | Deposit date: | 2017-12-22 | | Release date: | 2018-08-08 | | Last modified: | 2018-10-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural plasticity of the HHD2 domain of whirlin.
FEBS J., 285, 2018
|
|
4YGW
 
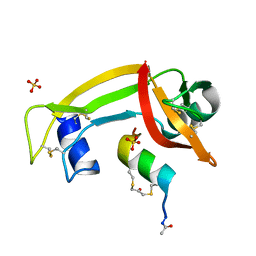 | | RNase S in complex with stabilized S peptide | | Descriptor: | 1-hydroxypropan-2-one, Ribonuclease A C2, S-peptide: ACE-LYS-GLU-THR-ALA-ALA-HCS-LYS-PHE-GLU-HCS-GLN-HIS-MET-ASP-SER, ... | | Authors: | Assem, N, Ferreira, D, Wolan, D.W, Dawson, P.E. | | Deposit date: | 2015-02-26 | | Release date: | 2015-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Acetone-Linked Peptides: A Convergent Approach for Peptide Macrocyclization and Labeling.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6FEJ
 
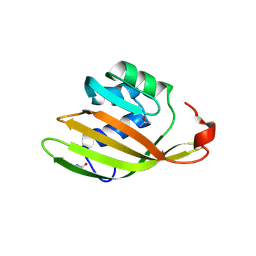 | | Anabaena Apo-C-Terminal Domain Homolog Protein | | Descriptor: | All4940 protein, UREA | | Authors: | Harris, D, Wilson, A, Muzzopappa, F, Kirilovsky, D, Adir, N. | | Deposit date: | 2018-01-02 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural rearrangements in the C-terminal domain homolog of Orange Carotenoid Protein are crucial for carotenoid transfer.
Commun Biol, 1, 2018
|
|
6FFB
 
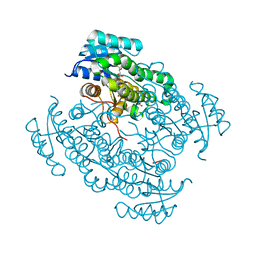 | | 17beta-hydroxysteroid dehydrogenase 14 variant S205 - mutant Q148A - in complex with a nonsteroidal inhibitor | | Descriptor: | 17-beta-hydroxysteroid dehydrogenase 14, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, ... | | Authors: | Bertoletti, N, Heine, A, Klebe, G, Marchais-Oberwinkler, S. | | Deposit date: | 2018-01-05 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mutational and structural studies uncover crucial amino acids determining activity and stability of 17 beta-HSD14.
J.Steroid Biochem.Mol.Biol., 189, 2019
|
|
8JFV
 
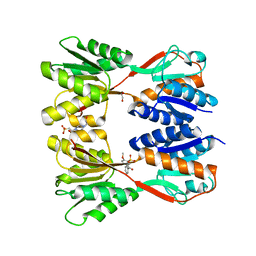 | | Crystal structure of Catabolite repressor acivator from E. coli in complex with sulisobenzone | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-4-oxidanyl-5-(phenylcarbonyl)benzenesulfonic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Neetu, N, Sharma, M, Mahto, J.K, Kumar, P. | | Deposit date: | 2023-05-19 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Sulisobenzone is a potent inhibitor of the global transcription factor Cra.
J.Struct.Biol., 215, 2023
|
|
7QBS
 
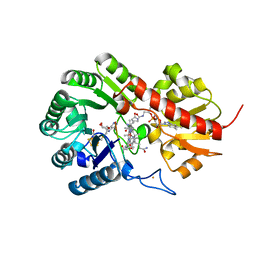 | | B12-dependent radical SAM methyltransferase, Mmp10 with [4Fe-4S] cluster, cobalamin, S-adenosyl-L-methionine, and peptide bound. | | Descriptor: | CO-METHYLCOBALAMIN, FE (III) ION, IRON/SULFUR CLUSTER, ... | | Authors: | Bernardo-Garcia, N, Fyfe, C.D, Chavas, L.M.G, Legrand, P, Benjdia, A, Berteau, O. | | Deposit date: | 2021-11-19 | | Release date: | 2022-02-02 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.327 Å) | | Cite: | Crystallographic snapshots of a B 12 -dependent radical SAM methyltransferase.
Nature, 602, 2022
|
|
4YFA
 
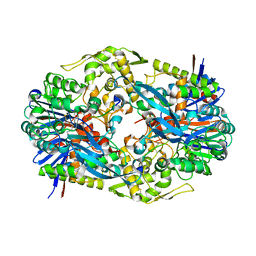 | |
6FDH
 
 | |
7QGI
 
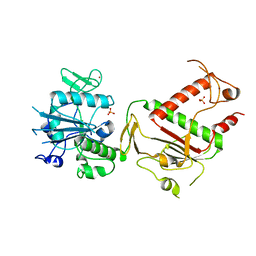 | | Crystal structure of SARS-CoV-2 NSP14 in the absence of NSP10 | | Descriptor: | PHOSPHATE ION, Proofreading exoribonuclease nsp14, ZINC ION | | Authors: | Newman, J.A, Imprachim, N, Yosaatmadja, Y, Gileadi, O. | | Deposit date: | 2021-12-08 | | Release date: | 2022-01-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures and fragment screening of SARS-CoV-2 NSP14 reveal details of exoribonuclease activation and mRNA capping and provide starting points for antiviral drug development.
Nucleic Acids Res., 51, 2023
|
|
8JJ3
 
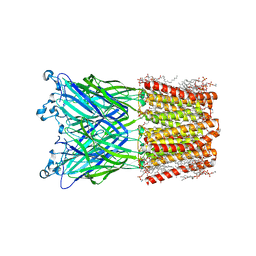 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 2.5 | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | Authors: | Bharambe, N, Li, Z, Basak, S. | | Deposit date: | 2023-05-29 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.6476 Å) | | Cite: | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
7QIF
 
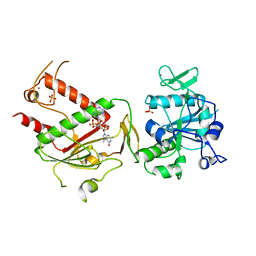 | | Crystal structure of SARS-CoV-2 NSP14 in complex with 7MeGpppG. | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, PHOSPHATE ION, Proofreading exoribonuclease nsp14, ... | | Authors: | Newman, J.A, Imprachim, N, Yosaatmadja, Y, Gileadi, O. | | Deposit date: | 2021-12-14 | | Release date: | 2022-02-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Crystal structures and fragment screening of SARS-CoV-2 NSP14 reveal details of exoribonuclease activation and mRNA capping and provide starting points for antiviral drug development.
Nucleic Acids Res., 51, 2023
|
|
4YS3
 
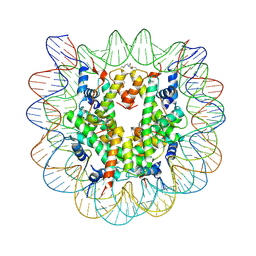 | | Nucleosome disassembly by RSC and SWI/SNF is enhanced by H3 acetylation near the nucleosome dyad axis | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Dechassa, M.L, Luger, K, Chatterjee, N, North, J.A, Manohar, M, Prasad, R, Ottessen, J.J, Poirier, M.G, Bartholomew, B. | | Deposit date: | 2015-03-16 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Histone Acetylation near the Nucleosome Dyad Axis Enhances Nucleosome Disassembly by RSC and SWI/SNF.
Mol.Cell.Biol., 35, 2015
|
|
8IYT
 
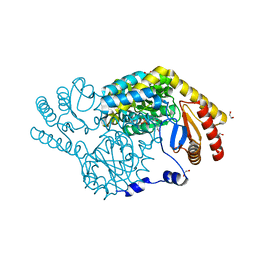 | | Crystal Structure of Serine Palmitoyltransferase complexed with D-methylserine | | Descriptor: | (2~{R})-2-methyl-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-3-oxidanyl-propanoic acid, 1,2-ETHANEDIOL, Serine palmitoyltransferase | | Authors: | Takahashi, A, Murakami, T, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2023-04-06 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Serine Palmitoyltransferase from Sphingobacterium multivorum
To Be Published
|
|
8J4G
 
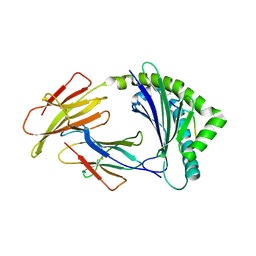 | | Crystal structure of 11JD mutant-I62N | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, peptide of AIV | | Authors: | Tang, Z, Zhang, N. | | Deposit date: | 2023-04-19 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure of duck MHC 11JD mutant-I62N
To Be Published
|
|
6FAQ
 
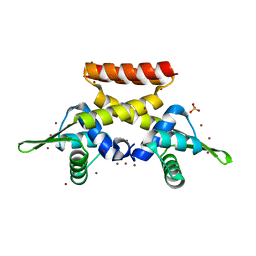 | |
8JFF
 
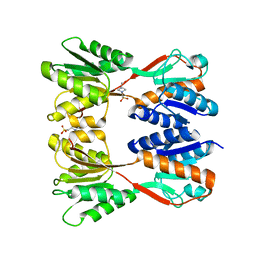 | |
8J2O
 
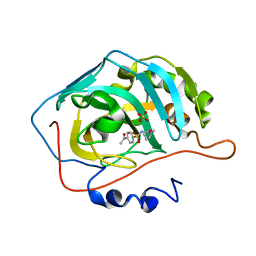 | |
8S9B
 
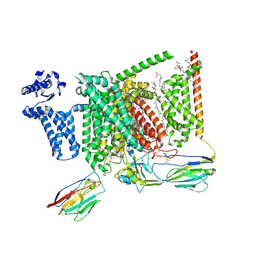 | | Cryo-EM structure of Nav1.7 with LCM | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2023-03-27 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
6F4I
 
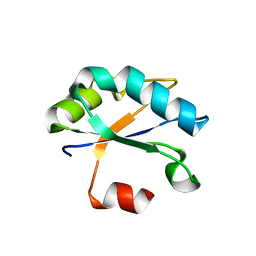 | | Crystal structure of Drosophila melanogaster SNF | | Descriptor: | U1 small nuclear ribonucleoprotein A | | Authors: | Weber, G, Holton, N, Hall, K.B, DeKoster, G, Wahl, M.C. | | Deposit date: | 2017-11-29 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Molecular principles underlying dual RNA specificity in the Drosophila SNF protein.
Nat Commun, 9, 2018
|
|
1WFQ
 
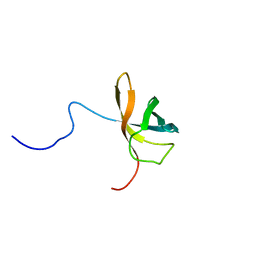 | | Solution structure of the first cold-shock domain of the human KIAA0885 protein (UNR protein) | | Descriptor: | UNR protein | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Tomizawa, T, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-26 | | Release date: | 2004-11-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
8S9C
 
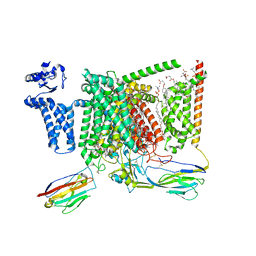 | | Cryo-EM structure of Nav1.7 with CBZ | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fan, X, Huang, J, Yan, N. | | Deposit date: | 2023-03-27 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
