5F3M
 
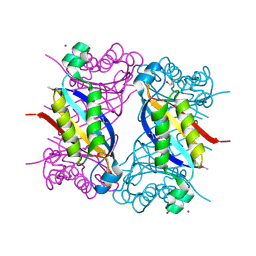 | | Crystal structure of dihydroneopterin aldolase from Bacillus anthracis complexed with L-neopterin at 1.5 Angstroms resolution . | | Descriptor: | 1,2-ETHANEDIOL, 7,8-dihydroneopterin aldolase, CHLORIDE ION, ... | | Authors: | Maltseva, N, Kim, Y, Shatsman, S, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-12-03 | | Release date: | 2015-12-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.498 Å) | | Cite: | Crystal structure of dihydroneopterin aldolase from Bacillus anthracis complexed with L-neopterin at 1.5 Angstroms resolution .
To Be Published
|
|
1SE0
 
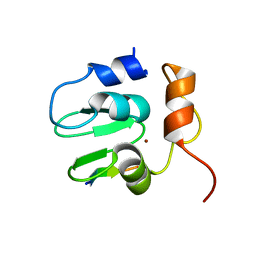 | | Crystal structure of DIAP1 BIR1 bound to a Grim peptide | | Descriptor: | Apoptosis 1 inhibitor, Cell death protein Grim, ZINC ION | | Authors: | Yan, N, Wu, J.W, Shi, Y. | | Deposit date: | 2004-02-15 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Molecular mechanisms of DrICE inhibition by DIAP1 and removal of inhibition by Reaper, Hid and Grim.
Nat.Struct.Mol.Biol., 11, 2004
|
|
6BYH
 
 | |
5F7A
 
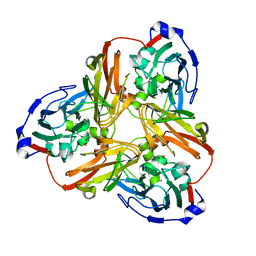 | | Nitrite complex structure of copper nitrite reductase from Alcaligenes faecalis determined at 293 K | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-12-07 | | Release date: | 2016-03-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1SEG
 
 | | Crystal structure of a toxin chimera between Lqh-alpha-IT from the scorpion Leiurus quinquestriatus hebraeus and AAH2 from Androctonus australis hector | | Descriptor: | AAH2: LQH-ALPHA-IT (FACE) CHIMERIC TOXIN, NITRATE ION, PROPANOIC ACID, ... | | Authors: | Karbat, I, Frolow, F, Froy, O, Gilles, N, Cohen, L, Turkov, M, Gordon, D, Gurevitz, M. | | Deposit date: | 2004-02-17 | | Release date: | 2004-08-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Molecular basis of the high insecticidal potency of scorpion alpha-toxins.
J.Biol.Chem., 279, 2004
|
|
5F5V
 
 | | Crystal structure of the Snu23-Prp38-MFAP1(217-296) complex of Chaetomium thermophilum | | Descriptor: | Prp38, Putative uncharacterized protein, Zinc finger domain-containing protein | | Authors: | Ulrich, A.K.C, Seeger, M, Bartlick, N, Wahl, M.C. | | Deposit date: | 2015-12-04 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Scaffolding in the Spliceosome via Single alpha Helices.
Structure, 24, 2016
|
|
5EVC
 
 | | Crystal structure of putative aspartate racemase from Salmonella Typhimurium complexed with sulfate and potassium | | Descriptor: | CHLORIDE ION, FLUORIDE ION, FORMIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Stam, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-19 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative aspartate racemase from Salmonella Typhimurium complexed with sulfate and potassium
To be published
|
|
5CQ3
 
 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with 6-Hydroxypicolinic acid (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 6-hydroxypyridine-2-carboxylic acid, Bromodomain adjacent to zinc finger domain protein 2B, ... | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-21 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with 6-Hydroxypicolinic acid (SGC - Diamond I04-1 fragment screening)
To be published
|
|
1SD7
 
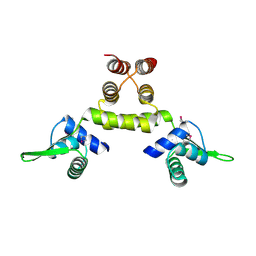 | | Crystal Structure of a SeMet derivative of MecI at 2.65 A | | Descriptor: | Methicillin resistance regulatory protein mecI | | Authors: | Safo, M.K, Zhao, Q, Musayev, F.N, Robinson, H, Scarsdale, N, Archer, G.L. | | Deposit date: | 2004-02-13 | | Release date: | 2004-02-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structures of the BlaI repressor from Staphylococcus aureus and its complex with DNA: insights into transcriptional regulation of the bla and mec operons
J.Bacteriol., 187, 2005
|
|
5CQQ
 
 | | Crystal structure of the Drosophila Zeste DNA binding domain in complex with DNA | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*AP*CP*GP*AP*GP*TP*GP*GP*AP*AP*AP*AP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*TP*TP*TP*TP*CP*CP*AP*CP*TP*CP*GP*TP*TP*TP*TP*T)-3'), Regulatory protein zeste | | Authors: | Gao, G.N, Wang, M, Yang, N, Huang, Y, Xu, R.M. | | Deposit date: | 2015-07-22 | | Release date: | 2015-11-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of Zeste-DNA Complex Reveals a New Modality of DNA Recognition by Homeodomain-Like Proteins
J.Mol.Biol., 427, 2015
|
|
1B3U
 
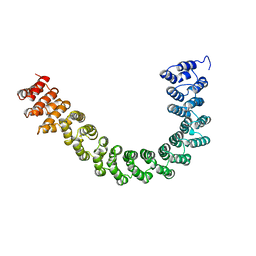 | | CRYSTAL STRUCTURE OF CONSTANT REGULATORY DOMAIN OF HUMAN PP2A, PR65ALPHA | | Descriptor: | PROTEIN (PROTEIN PHOSPHATASE PP2A) | | Authors: | Groves, M.R, Hanlon, N, Turowski, P, Hemmings, B, Barford, D. | | Deposit date: | 1998-12-14 | | Release date: | 1999-04-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs.
Cell(Cambridge,Mass.), 96, 1999
|
|
5CUD
 
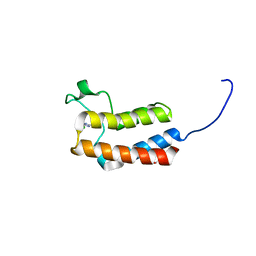 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with 6-CHLOROPURINE (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 6-chloro-9H-purine, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with 6-CHLOROPURINE (SGC - Diamond I04-1 fragment screening)
To be published
|
|
1B2V
 
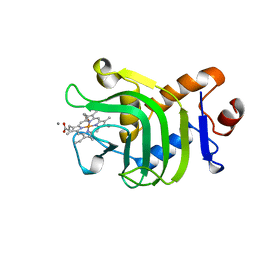 | | HEME-BINDING PROTEIN A | | Descriptor: | CALCIUM ION, PROTEIN (HEME-BINDING PROTEIN A), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arnoux, P, Haser, R, Izadi, N, Lecroisey, A, Wandersma, N.C, Czjzek, M. | | Deposit date: | 1998-12-01 | | Release date: | 1999-06-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of HasA, a hemophore secreted by Serratia marcescens.
Nat.Struct.Biol., 6, 1999
|
|
6BZW
 
 | | Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the GL precursor of the broadly neutralizing antibody AP33 | | Descriptor: | AP33 GL Heavy Chain, AP33 GL Light Chain, E2 AS412 peptide | | Authors: | Tzarum, N, Aleman, F, Wilson, I.A, Law, M. | | Deposit date: | 2017-12-26 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Immunogenetic and structural analysis of a class of HCV broadly neutralizing antibodies and their precursors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5DEA
 
 | | Crystal structure of the complex between human FMRP RGG motif and G-quadruplex RNA, cesium bound form. | | Descriptor: | CESIUM ION, Fragile X mental retardation protein 1, POTASSIUM ION, ... | | Authors: | Vasilyev, N, Polonskaia, A, Darnell, J.C, Darnell, R.B, Patel, D.J, Serganov, A. | | Deposit date: | 2015-08-25 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7973 Å) | | Cite: | Crystal structure reveals specific recognition of a G-quadruplex RNA by a beta-turn in the RGG motif of FMRP.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5D4H
 
 | | High-resolution nitrite complex of a copper nitrite reductase determined by synchrotron radiation crystallography | | Descriptor: | ACETIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1B70
 
 | | PHENYLALANYL TRNA SYNTHETASE COMPLEXED WITH PHENYLALANINE | | Descriptor: | MAGNESIUM ION, PHENYLALANINE, PHENYLALANYL-TRNA SYNTHETASE | | Authors: | Reshetnikova, L, Moor, N, Lavrik, O, Vassylyev, D.G. | | Deposit date: | 1999-01-26 | | Release date: | 2000-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of phenylalanyl-tRNA synthetase complexed with phenylalanine and a phenylalanyl-adenylate analogue
J.Mol.Biol., 287, 1999
|
|
1SSK
 
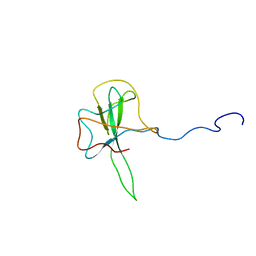 | | Structure of the N-terminal RNA-binding Domain of the SARS CoV Nucleocapsid Protein | | Descriptor: | Nucleocapsid protein | | Authors: | Huang, Q, Yu, L, Petros, A.M, Gunasekera, A, Liu, Z, Xu, N, Hajduk, P, Mack, J, Fesik, S.W, Olejniczak, E.T. | | Deposit date: | 2004-03-24 | | Release date: | 2004-06-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-Terminal RNA-Binding Domain of the SARS CoV Nucleocapsid Protein.
Biochemistry, 43, 2004
|
|
5DG6
 
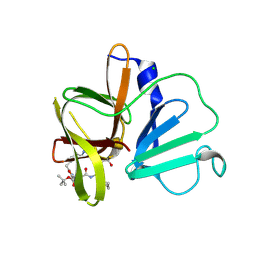 | | 2.35A resolution structure of Norovirus 3CL protease in complex an oxadiazole-based, cell permeable macrocyclic (21-mer) inhibitor | | Descriptor: | 3C-LIKE PROTEASE, CHLORIDE ION, tert-butyl [(4S,7S,10S)-7-(cyclohexylmethyl)-10-(hydroxymethyl)-5,8,13-trioxo-23-oxa-6,9,14,21,22-pentaazabicyclo[18.2.1]tricosa-1(22),20-dien-4-yl]carbamate | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Damalanka, V.C, Kim, Y, Alliston, K.R, Weerawarna, P.M, Kankanamalage, A.C.G, Lushington, G.H, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2015-08-27 | | Release date: | 2016-02-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Oxadiazole-Based Cell Permeable Macrocyclic Transition State Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 59, 2016
|
|
3L4G
 
 | | Crystal structure of Homo Sapiens cytoplasmic Phenylalanyl-tRNA synthetase | | Descriptor: | PHENYLALANINE, Phenylalanyl-tRNA synthetase alpha chain, Phenylalanyl-tRNA synthetase beta chain | | Authors: | Finarov, I, Moor, N, Kessler, N, Klipcan, L, Safro, M.G. | | Deposit date: | 2009-12-20 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of human cytosolic phenylalanyl-tRNA synthetase: evidence for kingdom-specific design of the active sites and tRNA binding patterns.
Structure, 18, 2010
|
|
5D9C
 
 | | Luciferin-regenerating enzyme solved by SIRAS using XFEL (refined against Hg derivative data) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Luciferin regenerating enzyme, MAGNESIUM ION, ... | | Authors: | Yamashita, K, Pan, D, Okuda, T, Murai, T, Kodan, A, Yamaguchi, T, Gomi, K, Kajiyama, N, Kato, H, Ago, H, Yamamoto, M, Nakatsu, T. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An isomorphous replacement method for efficient de novo phasing for serial femtosecond crystallography.
Sci Rep, 5, 2015
|
|
1BFW
 
 | | RETRO-INVERSO ANALOGUE OF THE G-H LOOP OF VP1 IN FOOT-AND-MOUTH-DISEASE (FMD) VIRUS, NMR, 10 STRUCTURES | | Descriptor: | VP1 PROTEIN | | Authors: | Petit, M.C, Benkirane, N, Guichard, G, Phan Chan Du, A, Cung, M.T, Briand, J.P, Muller, S. | | Deposit date: | 1998-05-22 | | Release date: | 1999-01-13 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a retro-inverso peptide analogue mimicking the foot-and-mouth disease virus major antigenic site. Structural basis for its antigenic cross-reactivity with the parent peptide.
J.Biol.Chem., 274, 1999
|
|
1SV9
 
 | | Crystal structure of the complex formed between groupII phospholipase A2 and anti-inflammatory agent 2-[(2,6-Dichlorophenyl)amino] benzeneacetic acid at 2.7A resolution | | Descriptor: | 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, Phospholipase A2 | | Authors: | Senthil kumar, R, Singh, N, Ethayathulla, A.S, Prem kumar, R, Sharma, S, Singh, T.P. | | Deposit date: | 2004-03-29 | | Release date: | 2004-04-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Crystal structure of the complex formed between group II phospholipase A2 and anti-inflammatory agent 2-[(2,6-Dichlorophenyl)amino] benzeneacetic acid at 2.7A resolution
To be Published
|
|
2WZZ
 
 | |
5DHH
 
 | | The crystal structure of nociceptin/orphanin FQ peptide receptor (NOP) in complex with SB-612111 (PSI Community Target) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (5S,7S)-7-{[4-(2,6-dichlorophenyl)piperidin-1-yl]methyl}-1-methyl-6,7,8,9-tetrahydro-5H-benzo[7]annulen-5-ol, OLEIC ACID, ... | | Authors: | Miller, R.L, Thompson, A.A, Trapella, C, Guerrini, R, Malfacini, D, Patel, N, Han, G.W, Cherezov, V, Calo, G, Katritch, V, Stevens, R.C, GPCR Network (GPCR) | | Deposit date: | 2015-08-31 | | Release date: | 2015-10-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | The Importance of Ligand-Receptor Conformational Pairs in Stabilization: Spotlight on the N/OFQ G Protein-Coupled Receptor.
Structure, 23, 2015
|
|
