1OXX
 
 | | Crystal structure of GlcV, the ABC-ATPase of the glucose ABC transporter from Sulfolobus solfataricus | | Descriptor: | ABC transporter, ATP binding protein, IODIDE ION | | Authors: | Verdon, G, Albers, S.-V, van Oosterwijk, N, Dijkstra, B.W, Driessen, A.J.M, Thunnissen, A.M.W.H. | | Deposit date: | 2003-04-03 | | Release date: | 2003-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Formation of the productive ATP-Mg2+-bound dimer of GlcV, an ABC-ATPase from Sulfolobus solfataricus
J.Mol.Biol., 334, 2003
|
|
6P6W
 
 | | Cryo-EM structure of voltage-gated sodium channel NavAb N49K/L109A/M116V/G94C/Q150C disulfide crosslinked mutant in the resting state | | Descriptor: | Fusion of Maltose-binding protein and voltage-gated sodium channel NavAb | | Authors: | Wisedchaisri, G, Tonggu, L, McCord, E, Gamal El-Din, T.M, Wang, L, Zheng, N, Catterall, W.A. | | Deposit date: | 2019-06-04 | | Release date: | 2019-08-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Resting-State Structure and Gating Mechanism of a Voltage-Gated Sodium Channel.
Cell, 178, 2019
|
|
5CBM
 
 | | Crystal structure of PfA-M17 with virtual ligand inhibitor | | Descriptor: | (2S)-2-{[(R)-[(R)-amino(phenyl)methyl](hydroxy)phosphoryl]methyl}-4-methylpentanoic acid, CARBONATE ION, GLYCEROL, ... | | Authors: | Ruggeri, C, Drinkwater, N, McGowan, S. | | Deposit date: | 2015-07-01 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and Validation of a Potent Dual Inhibitor of the P. falciparum M1 and M17 Aminopeptidases Using Virtual Screening.
Plos One, 10, 2015
|
|
6UE7
 
 | | Structure of dimeric sIgA complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, ... | | Authors: | Kumar, N, Arthur, C.P, Ciferri, C, Matsumoto, M.L. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the secretory immunoglobulin A core.
Science, 367, 2020
|
|
6U0Y
 
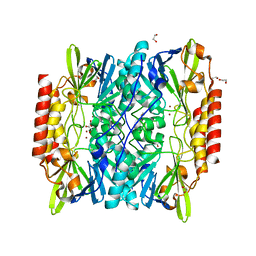 | | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia | | Descriptor: | 1,2-ETHANEDIOL, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ZINC ION | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-08-15 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the metallo-beta-lactamase L1 from Stenotrophomonas maltophilia
To Be Published
|
|
1OIB
 
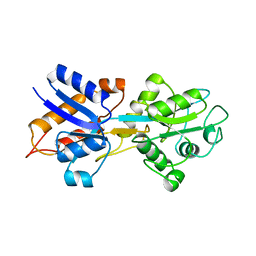 | |
6U2D
 
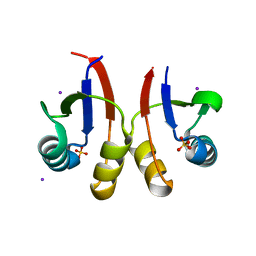 | | PmtCD peptide exporter basket domain | | Descriptor: | ABC transporter ATP-binding protein, IODIDE ION, SULFATE ION | | Authors: | Zeytuni, N, Strynadka, N.C.J. | | Deposit date: | 2019-08-19 | | Release date: | 2020-10-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural insight into the Staphylococcus aureus ATP-driven exporter of virulent peptide toxins
Sci Adv, 6, 2020
|
|
6P2J
 
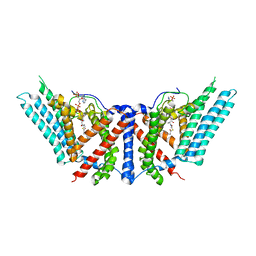 | | Dimeric structure of ACAT1 | | Descriptor: | S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (9Z)-octadec-9-enethioate (non-preferred name), Sterol O-acyltransferase 1 | | Authors: | Yan, N, Qian, H.W. | | Deposit date: | 2019-05-21 | | Release date: | 2020-05-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for catalysis and substrate specificity of human ACAT1.
Nature, 581, 2020
|
|
6UEA
 
 | | Structure of pentameric sIgA complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, ... | | Authors: | Kumar, N, Arthur, C.P, Ciferri, C, Matsumoto, M.L. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the secretory immunoglobulin A core.
Science, 367, 2020
|
|
5CNN
 
 | | Crystal structure of the EGFR kinase domain mutant I682Q | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Kovacs, E, Das, R, Mirza, A, Jura, N, Barros, T, Kuriyan, J. | | Deposit date: | 2015-07-17 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analysis of the Role of the C-Terminal Tail in the Regulation of the Epidermal Growth Factor Receptor.
Mol.Cell.Biol., 35, 2015
|
|
1OGP
 
 | | The crystal structure of plant sulfite oxidase provides insight into sulfite oxidation in plants and animals | | Descriptor: | (MOLYBDOPTERIN-S,S)-DIOXO-THIO-MOLYBDENUM(VI), CESIUM ION, GLYCEROL, ... | | Authors: | Schrader, N, Fischer, K, Theis, K, Mendel, R.R, Schwarz, G, Kisker, C. | | Deposit date: | 2003-05-08 | | Release date: | 2003-10-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of Plant Sulfite Oxidase Provides Insights Into Sulfite Oxidation in Plants and Animals
Structure, 11, 2003
|
|
6FBR
 
 | | Crystal Structure of the Human Retinoid X Receptor DNA-Binding Domain Bound to the Human MEp DR1 Response Element, pH 4.2 | | Descriptor: | DI(HYDROXYETHYL)ETHER, DNA (5'-D(*CP*TP*GP*GP*GP*TP*CP*AP*AP*AP*GP*TP*TP*CP*AP*TP*C)-3'), DNA (5'-D(*GP*AP*TP*GP*AP*AP*CP*TP*TP*TP*GP*AP*CP*CP*CP*AP*G)-3'), ... | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Osz, J, Rochel, N. | | Deposit date: | 2017-12-19 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Modulation of RXR-DNA complex assembly by DNA context.
Mol. Cell. Endocrinol., 481, 2019
|
|
6TVE
 
 | | Unliganded human CD73 (5'-nucleotidase) in the open state | | Descriptor: | 5'-nucleotidase, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Scaletti, E, Strater, N. | | Deposit date: | 2020-01-09 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | 2-Substituted alpha , beta-Methylene-ADP Derivatives: Potent Competitive Ecto-5'-nucleotidase (CD73) Inhibitors with Variable Binding Modes.
J.Med.Chem., 63, 2020
|
|
5CD9
 
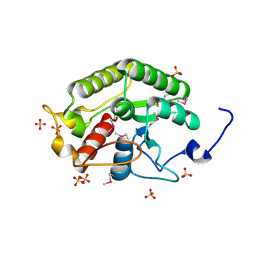 | | Crystal structure of the CTD of Drosophila Oskar protein | | Descriptor: | Maternal effect protein oskar, SULFATE ION | | Authors: | Yang, N, Hu, M, Yu, Z, Wang, M, Lehmann, R, Xu, R.M. | | Deposit date: | 2015-07-03 | | Release date: | 2015-09-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structure of Drosophila Oskar reveals a novel RNA binding protein
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6CMO
 
 | | Rhodopsin-Gi complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab Heavy chain, Fab light chain, ... | | Authors: | Kang, Y, Kuybeda, O, de Waal, P.W, Mukherjee, S, Van Eps, N, Dutka, P, Zhou, X.E, Bartesaghi, A, Erramilli, S, Morizumi, T, Gu, X, Yin, Y, Liu, P, Jiang, Y, Meng, X, Zhao, G, Melcher, K, Earnst, O.P, Kossiakoff, A.A, Subramaniam, S, Xu, H.E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-06-20 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of human rhodopsin bound to an inhibitory G protein.
Nature, 558, 2018
|
|
6TZL
 
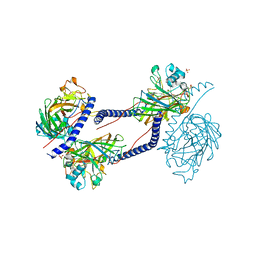 | |
6CO9
 
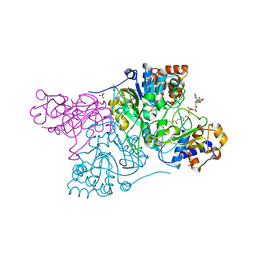 | | Crystal structure of Rhodococcus jostii RHA1 IpdAB COCHEA-COA complex | | Descriptor: | Probable CoA-transferase alpha subunit, Probable CoA-transferase beta subunit, S-{(3R,5R,9R)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-3,5-dioxido-10,14-dioxo-2,4,6-trioxa-11,15-diaza-3lambda~5~,5lambda~5~-diphosphaheptadecan-17-yl} (5R,10R)-7-hydroxy-10-methyl-2-oxo-1-oxaspiro[4.5]dec-6-ene-6-carbothioate (non-preferred name), ... | | Authors: | Crowe, A.M, Workman, S.D, Watanabe, N, Worrall, L.J, Strynadka, N.C.J, Eltis, L.D. | | Deposit date: | 2018-03-12 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | IpdAB, a virulence factor inMycobacterium tuberculosis, is a cholesterol ring-cleaving hydrolase.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5CEE
 
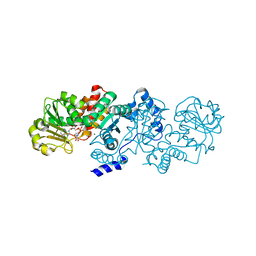 | | Malic enzyme from Candidatus Phytoplasma AYWB in complex with NAD and Mg2+ | | Descriptor: | MAGNESIUM ION, NAD-dependent malic enzyme, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Alvarez, C.E, Trajtenberg, F, Larrieux, N, Saigo, M, Mussi, M.A, Andreo, C.S, Drincovich, M.F, Buschiazzo, A. | | Deposit date: | 2015-07-06 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | The crystal structure of the malic enzyme from Candidatus Phytoplasma reveals the minimal structural determinants for a malic enzyme.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
1QG0
 
 | | WILD-TYPE PEA FNR | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PROTEIN (FERREDOXIN:NADP+ REDUCTASE) | | Authors: | Deng, Z, Aliverti, A, Zanetti, G, Arakaki, A.K, Ottado, J, Orellano, E.G, Calcaterra, N.B, Ceccarelli, E.A, Carrillo, N, Karplus, P.A. | | Deposit date: | 1999-04-18 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A productive NADP+ binding mode of ferredoxin-NADP+ reductase revealed by protein engineering and crystallographic studies.
Nat.Struct.Biol., 6, 1999
|
|
1QGA
 
 | | PEA FNR Y308W MUTANT IN COMPLEX WITH NADP+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTEIN (FERREDOXIN:NADP+ REDUCTASE), ... | | Authors: | Deng, Z, Aliverti, A, Zanetti, G, Arakaki, A.K, Ottado, J, Orellano, E.G, Calcaterra, N.B, Ceccarelli, E.A, Carrillo, N, Karplus, P.A. | | Deposit date: | 1999-04-18 | | Release date: | 1999-04-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A productive NADP+ binding mode of ferredoxin-NADP+ reductase revealed by protein engineering and crystallographic studies.
Nat.Struct.Biol., 6, 1999
|
|
6FIR
 
 | |
6P1I
 
 | | Structure of HIV-1 Reverse Transcriptase (RT) in complex with dsDNA and dCTP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, DNA Primer 20-mer, DNA template 27-mer, ... | | Authors: | Bertoletti, N, Anderson, K.S. | | Deposit date: | 2019-05-19 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural insights into the recognition of nucleoside reverse transcriptase inhibitors by HIV-1 reverse transcriptase: First crystal structures with reverse transcriptase and the active triphosphate forms of lamivudine and emtricitabine.
Protein Sci., 28, 2019
|
|
7ZEM
 
 | |
7ZEO
 
 | |
6FLJ
 
 | |
