6O78
 
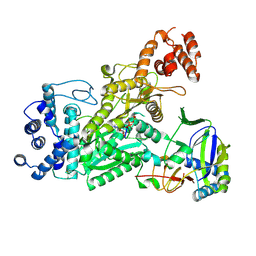 | | Crystal structure of Csm1-Csm4 cassette in complex with pppApApA | | Descriptor: | CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), Csm4, MANGANESE (II) ION, ... | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Second Messenger cA4Formation within the Composite Csm1 Palm Pocket of Type III-A CRISPR-Cas Csm Complex and Its Release Path.
Mol.Cell, 75, 2019
|
|
6OU9
 
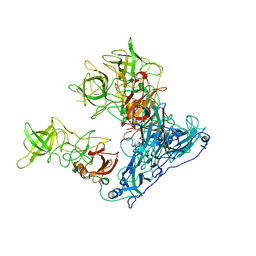 | | Asymmetric focused reconstruction of human norovirus GI.7 Houston strain VLP asymmetric unit in T=3 symmetry | | Descriptor: | Major capsid protein | | Authors: | Jung, J, Grant, T, Thomas, D.R, Diehnelt, C.W, Grigorieff, N, Joshua-Tor, L. | | Deposit date: | 2019-05-04 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | High-resolution cryo-EM structures of outbreak strain human norovirus shells reveal size variations.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6O22
 
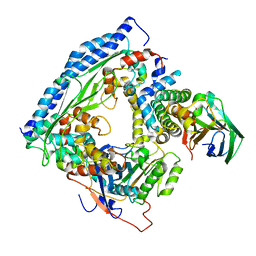 | | Structure of Asf1-H3:H4-Rtt109-Vps75 histone chaperone-lysine acetyltransferase complex with the histone substrate. | | Descriptor: | Histone H3.2, Histone H4, Histone acetyltransferase RTT109, ... | | Authors: | Danilenko, N, Carlomagno, T, Kirkpatrick, J.P. | | Deposit date: | 2019-02-22 | | Release date: | 2019-07-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR, SOLUTION SCATTERING | | Cite: | Histone chaperone exploits intrinsic disorder to switch acetylation specificity.
Nat Commun, 10, 2019
|
|
6NZ6
 
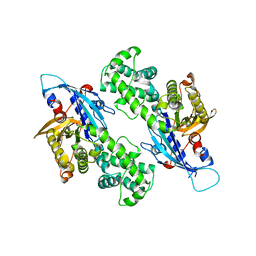 | | YcjX-GDP (type II) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, YcjX Stress Protein | | Authors: | Tsai, J, Sung, N, Lee, J, Chang, C, Lee, S, Tsai, F.T. | | Deposit date: | 2019-02-12 | | Release date: | 2019-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the YcjX Stress Protein Reveals a Ras-Like GTP-Binding Protein.
J.Mol.Biol., 431, 2019
|
|
6NY1
 
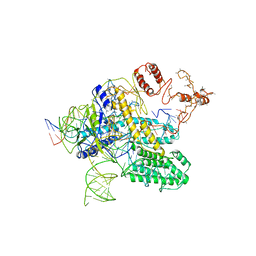 | | CasX-gRNA-DNA(30bp) State II | | Descriptor: | CasX, DNA Non-target strand, DNA target strand, ... | | Authors: | Liu, J.J, Orlova, N, Nogales, E, Doudna, J.A. | | Deposit date: | 2019-02-10 | | Release date: | 2019-02-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | CasX enzymes comprise a distinct family of RNA-guided genome editors.
Nature, 566, 2019
|
|
6OUT
 
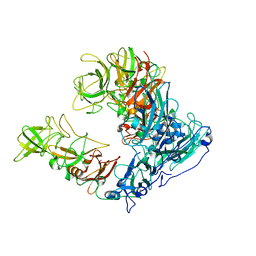 | | Asymmetric focused reconstruction of human norovirus GI.1 Norwalk strain VLP asymmetric unit in T=3 symmetry | | Descriptor: | Capsid protein VP1 | | Authors: | Jung, J, Grant, T, Thomas, D.R, Diehnelt, C.W, Grigorieff, N, Joshua-Tor, L. | | Deposit date: | 2019-05-05 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | High-resolution cryo-EM structures of outbreak strain human norovirus shells reveal size variations.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6OEG
 
 | | Structure of CagX from a cryo-EM reconstruction of a T4SS | | Descriptor: | Type IV secretion system apparatus protein CagX | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-27 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
6OVW
 
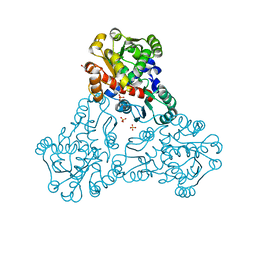 | | Crystal structure of ornithine carbamoyltransferase from Salmonella enterica | | Descriptor: | GLYCEROL, Ornithine carbamoyltransferase, PHOSPHATE ION | | Authors: | Chang, C, Mesa, N, Skarina, T, Savchenko, A, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-08 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.903 Å) | | Cite: | Crystal structure of ornithine carbamoyltransferase from Salmonella enterica
To Be Published
|
|
6O96
 
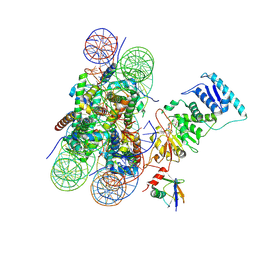 | | Dot1L bound to the H2BK120 Ubiquitinated nucleosome | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Valencia-Sanchez, M.I, De Ioannes, P.E, Miao, W, Vasilyev, N, Chen, R, Nudler, E, Armache, J.-P, Armache, K.-J. | | Deposit date: | 2019-03-13 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Basis of Dot1L Stimulation by Histone H2B Lysine 120 Ubiquitination.
Mol.Cell, 74, 2019
|
|
6ODJ
 
 | | PolyAla Model of the PRC from the Type 4 Secretion System of H. pylori | | Descriptor: | PolyAla Model of PRC from H.pylori | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-26 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
1YCD
 
 | | Crystal structure of yeast FSH1/YHR049W, a member of the serine hydrolase family | | Descriptor: | 2-HYDROXY-4,5-DIOXOHEPTYL HYDROGEN PHOSPHONATE, Hypothetical 27.3 kDa protein in AAP1-SMF2 intergenic region | | Authors: | Leulliot, N, Graille, M, Coste, F, Quevillon-Cheruel, S, Janin, J, van Tilbeurgh, H, Paris-Sud Yeast Structural Genomics (YSG) | | Deposit date: | 2004-12-22 | | Release date: | 2005-05-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of yeast YHR049W/FSH1, a member of the serine hydrolase family.
Protein Sci., 14, 2005
|
|
6N87
 
 | | Plasmodium falciparum FVO apical membrane antigen 1 (AMA1) bound to MTSL spin-labelled cyclised RON2 peptide | | Descriptor: | Apical membrane antigen-1, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate, backbone-cyclised peptide bcRON2hp | | Authors: | McGowan, S, Drinkwater, N. | | Deposit date: | 2018-11-28 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.588 Å) | | Cite: | Identification of the Binding Site of Apical Membrane Antigen 1 (AMA1) Inhibitors Using a Paramagnetic Probe.
ChemMedChem, 14, 2019
|
|
6N8T
 
 | | Hsp104DWB closed conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein 104 | | Authors: | Lee, S, Rho, S.H, Lee, J, Sung, N, Liu, J, Tsai, F.T.F. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Cryo-EM Structures of the Hsp104 Protein Disaggregase Captured in the ATP Conformation.
Cell Rep, 26, 2019
|
|
6NE3
 
 | | Cryo-EM structure of singly-bound SNF2h-nucleosome complex with SNF2h bound at SHL-2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (156-MER), Histone H2A type 1, ... | | Authors: | Armache, J.-P, Gamarra, N, Johnson, S.L, Leonard, J.D, Wu, S, Narlikar, G.N, Cheng, Y. | | Deposit date: | 2018-12-16 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structures of remodeler-nucleosome intermediates suggest allosteric control through the nucleosome.
Elife, 8, 2019
|
|
6ODB
 
 | | Crystal structure of HDAC8 in complex with compound 3 | | Descriptor: | GLYCEROL, Histone deacetylase 8, N-{2-[(1E)-3-(hydroxyamino)-3-oxoprop-1-en-1-yl]phenyl}-2-phenoxybenzamide, ... | | Authors: | Zheng, X, Conti, C, Caravella, J, Zablocki, M.-M, Bair, K, Barczak, N, Han, B, Lancia Jr, D, Liu, C, Martin, M, Ng, P.Y, Rudnitskaya, A, Thomason, J.J, Garcia-Dancey, R, Hardy, C, Lahdenranta, J, Leng, C, Li, P, Pardo, E, Saldahna, A, Tan, T, Toms, A.V, Yao, L, Zhang, C. | | Deposit date: | 2019-03-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
To Be Published
|
|
6OE3
 
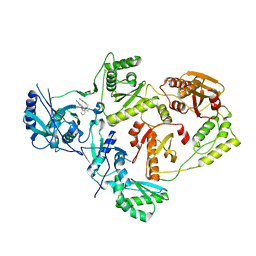 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 5-(2-(2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy)phenoxy)-7-fluoro-2-naphthonitrile (JLJ635), a Non-nucleoside Inhibitor | | Descriptor: | 5-{2-[2-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)ethoxy]phenoxy}-7-fluoronaphthalene-2-carbonitrile, HIV-1 REVERSE TRANSCRIPTASE, P51 SUBUNIT, ... | | Authors: | Bertoletti, N, Kudalkar, S.N, Anderson, K.S, Cisneros Trigo, J.A, Jorgensen, W.L. | | Deposit date: | 2019-03-27 | | Release date: | 2019-05-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and pharmacological evaluation of a novel non-nucleoside reverse transcriptase inhibitor as a promising long acting nanoformulation for treating HIV.
Antiviral Res., 167, 2019
|
|
6OEH
 
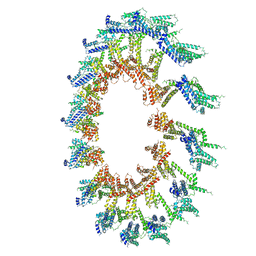 | | PolyAla Model of OMCC I-Layer | | Descriptor: | PolyAla Model of OMCC I-Layer | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-27 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
6OS4
 
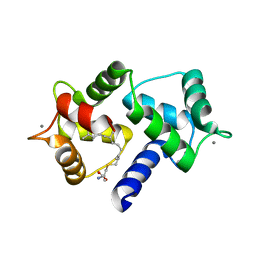 | | Calmodulin in complex with farnesyl cysteine methyl ester | | Descriptor: | CALCIUM ION, Calmodulin-1, s-farnesyl-l-cysteine methyl ester | | Authors: | Grant, B.M.M, Enomoto, M, Lee, K.Y, Back, S.I, Gebregiworgis, T, Ishiyama, N, Ikura, M, Marshall, C. | | Deposit date: | 2019-05-01 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Calmodulin disrupts plasma membrane localization of farnesylated KRAS4b by sequestering its lipid moiety.
Sci.Signal., 13, 2020
|
|
6OJ5
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (TLP_RNA) | | Descriptor: | Inner capsid protein VP2, RNA-directed RNA polymerase | | Authors: | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (5.2 Å) | | Cite: | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6O6V
 
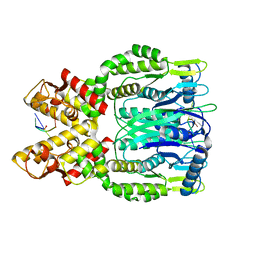 | |
6O73
 
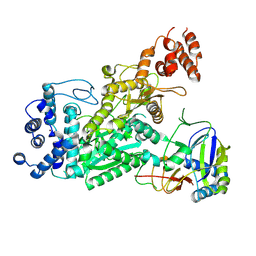 | | Crystal structure of apo Csm1-Csm4 cassette | | Descriptor: | Csm1, Csm4, NICKEL (II) ION | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Second Messenger cA4Formation within the Composite Csm1 Palm Pocket of Type III-A CRISPR-Cas Csm Complex and Its Release Path.
Mol.Cell, 75, 2019
|
|
6OEF
 
 | | PolyAla Model of the O-layer from the Type 4 Secretion System of H. pylori | | Descriptor: | PolyAla Model of OMCC O-Layer | | Authors: | Chung, J.M, Sheedlo, M.J, Campbell, A, Sawhney, N, Frick-Cheng, A.E, Lacy, D.B, Cover, T.L, Ohi, M.D. | | Deposit date: | 2019-03-27 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the Helicobacter pylori Cag type IV secretion system.
Elife, 8, 2019
|
|
6O70
 
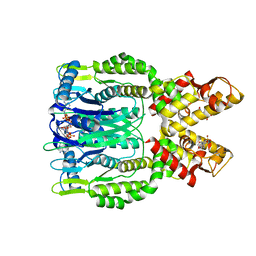 | | Crystal structure of Csm6 H132A mutant in complex with cA4 by cocrystallization of cA4 and Csm6 H132A mutant | | Descriptor: | 2',3'- cyclic AMP, 3'-O-[(R)-{[(2S,3aS,4S,6S,6aS)-6-(6-amino-9H-purin-9-yl)-2-hydroxy-2-oxotetrahydro-2H-2lambda~5~-furo[3,4-d][1,3,2]dioxaphosphol-4-yl]methoxy}(hydroxy)phosphoryl]adenosine, Csm6 | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-03-07 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | CRISPR-Cas III-A Csm6 CARF Domain Is a Ring Nuclease Triggering Stepwise cA4Cleavage with ApA>p Formation Terminating RNase Activity.
Mol.Cell, 75, 2019
|
|
6OJ4
 
 | | In situ structure of rotavirus VP1 RNA-dependent RNA polymerase (DLP) | | Descriptor: | Inner capsid protein VP2, RNA-directed RNA polymerase | | Authors: | Jenni, S, Salgado, E.N, Herrmann, T, Li, Z, Grant, T, Grigorieff, N, Trapani, S, Estrozi, L.F, Harrison, S.C. | | Deposit date: | 2019-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In situ Structure of Rotavirus VP1 RNA-Dependent RNA Polymerase.
J.Mol.Biol., 431, 2019
|
|
6ZLO
 
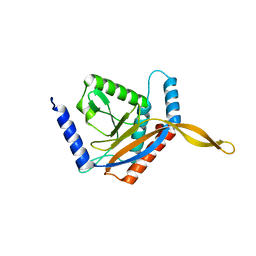 | | E2 core of the fungal Pyruvate dehydrogenase complex with asymmetric interior PX30 component | | Descriptor: | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial | | Authors: | Forsberg, B.O, Howard, R.J, Aibara, S, Mortesaei, N, Lindahl, E. | | Deposit date: | 2020-06-30 | | Release date: | 2020-09-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Arrangement and symmetry of the fungal E3BP-containing core of the pyruvate dehydrogenase complex.
Nat Commun, 11, 2020
|
|
