1MDF
 
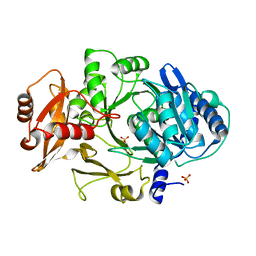 | | CRYSTAL STRUCTURE OF DhbE IN ABSENCE OF SUBSTRATE | | Descriptor: | 2,3-dihydroxybenzoate-AMP ligase, SULFATE ION | | Authors: | May, J.J, Kessler, N, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2002-08-07 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1MA8
 
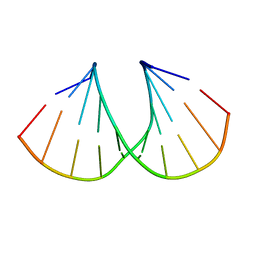 | | A-DNA decamer GCGTA(UMS)ACGC with incorporated 2'-methylseleno-uridine | | Descriptor: | 5'-D(*GP*CP*GP*TP*AP*UMSP*AP*CP*GP*C)-3' | | Authors: | Teplova, M, Wilds, C.J, Wawrzak, Z, Tereshko, V, Du, Q, Carrasco, N, Huang, Z, Egli, M. | | Deposit date: | 2002-08-01 | | Release date: | 2002-12-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Covalent incorporation of selenium into oligonucleotides for X-ray crystal structure determination via MAD: proof of principle
BIOCHIMIE, 84, 2002
|
|
6N7W
 
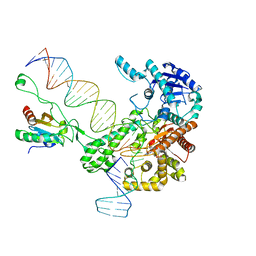 | | Structure of bacteriophage T7 leading-strand DNA polymerase (D5A/E7A)/Trx in complex with a DNA fork and incoming dTTP (from multiple lead complexes) | | Descriptor: | DNA (25-MER), DNA (77-MER), DNA-directed DNA polymerase, ... | | Authors: | Gao, Y, Fox, T, Val, N, Yang, W. | | Deposit date: | 2018-11-28 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
1M4W
 
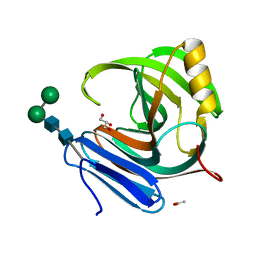 | | Thermophilic b-1,4-xylanase from Nonomuraea flexuosa | | Descriptor: | ACETATE ION, GLYCEROL, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hakulinen, N, Turunen, O, Janis, J, Leisola, M, Rouvinen, J. | | Deposit date: | 2002-07-05 | | Release date: | 2003-07-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structures of thermophilic beta-1,4-xylanases from Chaetomium thermophilum and Nonomuraea flexuosa. Comparison of twelve xylanases in relation to their thermal stability.
Eur.J.Biochem., 270, 2003
|
|
1M53
 
 | |
8BQ6
 
 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Complete conformation 2 composition) | | Descriptor: | 2,3-DIMETHOXY-5-METHYL-6-(3,11,15,19-TETRAMETHYL-EICOSA-2,6,10,14,18-PENTAENYL)-[1,4]BENZOQUINONE, AT3G07480.1, Acyl carrier protein 1, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-11-18 | | Release date: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
1MH2
 
 | | Crystal Structure of a Zinc Containing Dimer of Phospholipase A2 from the Venom of Indian Cobra (Naja Naja Sagittifera) | | Descriptor: | ACETIC ACID, PHOSPHOLIPASE A2, ZINC ION | | Authors: | Jabeen, T, Varma, A.K, Paramasivam, M, Singh, N, Singh, R.K, Sharma, S, Srinivasan, A, Singh, T.P. | | Deposit date: | 2002-08-19 | | Release date: | 2003-05-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of a Zinc Containing Dimer of Phospholipase A2 from the Venom of Indian cobra (Naja Naja Saggittifera)
To be Published
|
|
1MDB
 
 | | CRYSTAL STRUCTURE OF DhbE IN COMPLEX WITH DHB-ADENYLATE | | Descriptor: | 2,3-DIHYDROXY-BENZOIC ACID, 2,3-dihydroxybenzoate-AMP ligase, ADENOSINE MONOPHOSPHATE, ... | | Authors: | May, J.J, Kessler, N, Marahiel, M.A, Stubbs, M.T. | | Deposit date: | 2002-08-07 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1MHY
 
 | | METHANE MONOOXYGENASE HYDROXYLASE | | Descriptor: | FE (III) ION, METHANE MONOOXYGENASE HYDROXYLASE | | Authors: | Elango, N, Radhakrishnan, R, Froland, W.A, Waller, B.J, Earhart, C.A, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1996-10-21 | | Release date: | 1997-05-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the hydroxylase component of methane monooxygenase from Methylosinus trichosporium OB3b
Protein Sci., 6, 1997
|
|
1MQB
 
 | | Crystal Structure of Ephrin A2 (ephA2) Receptor Protein Kinase | | Descriptor: | Ephrin type-A receptor 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Nowakowski, J, Cronin, C.N, McRee, D.E, Knuth, M.W, Nelson, C, Pavletich, N, Rogers, J, Sang, B.C, Scheibe, D.N, Swanson, R.V, Thompson, D.A. | | Deposit date: | 2002-09-16 | | Release date: | 2003-09-16 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the Cancer Related Aurora-A, FAK and EphA2 Protein Kinases from Nanovolume Crystallography
Structure, 10, 2003
|
|
1MPV
 
 | | Structure of bhpBR3, the BAFF-binding loop of BR3 embedded in a beta-hairpin peptide | | Descriptor: | BLyS Receptor 3 | | Authors: | Kayagaki, N, Yan, M, Seshasayee, D, Wang, H, Lee, W, French, D.M, Grewal, I.S, Cochran, A.G, Gordon, N.C, Yin, J, Starovasnik, M.A, Dixit, V.M. | | Deposit date: | 2002-09-12 | | Release date: | 2002-10-30 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | BAFF/BLyS receptor 3 binds the B cell survival factor BAFF ligand through a discrete surface loop and promotes processing of NF-kappaB2.
Immunity, 17, 2002
|
|
1MFS
 
 | | DYNAMICAL BEHAVIOR OF THE HIV-1 NUCLEOCAPSID PROTEIN; NMR, 30 STRUCTURES | | Descriptor: | HIV-1 NUCLEOCAPSID PROTEIN, ZINC ION | | Authors: | Summers, M.F, Turner, B.G, De Guzman, R.N, Lee, B.M, Tjandra, N. | | Deposit date: | 1998-04-01 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Dynamical behavior of the HIV-1 nucleocapsid protein.
J.Mol.Biol., 279, 1998
|
|
1MH8
 
 | | Crystal Structure of a Phopholipase A2 Monomer with Isoleucine at Second Position | | Descriptor: | PHOSPHOLIPASE A2 | | Authors: | Jabeen, T, Jasti, J, Singh, N, Singh, R.K, Sharma, S, Singh, T.P. | | Deposit date: | 2002-08-19 | | Release date: | 2003-06-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal Structure of a Phospholipase A2 Monomer with Isoleucine at Second Position
To be Published
|
|
1COO
 
 | | THE COOH-TERMINAL DOMAIN OF RNA POLYMERASE ALPHA SUBUNIT | | Descriptor: | RNA POLYMERASE ALPHA SUBUNIT | | Authors: | Jeon, Y.H, Negishi, T, Shirakawa, M, Yamazaki, T, Fujita, N, Ishihama, A, Kyogoku, Y. | | Deposit date: | 1995-10-09 | | Release date: | 1996-03-08 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the activator contact domain of the RNA polymerase alpha subunit.
Science, 270, 1995
|
|
1OXF
 
 | | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins | | Descriptor: | cyan fluorescent protein cfp | | Authors: | Hyun Bae, J, Rubini, M, Jung, G, Wiegand, G, Seifert, M.H, Azim, M.K, Kim, J.S, Zumbusch, A, Holak, T.A, Moroder, L, Huber, R, Budisa, N. | | Deposit date: | 2003-04-02 | | Release date: | 2003-12-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins
J.Mol.Biol., 328, 2003
|
|
1OVL
 
 | | Crystal Structure of Nurr1 LBD | | Descriptor: | BROMIDE ION, IODIDE ION, Orphan nuclear receptor NURR1 (MSE 414, ... | | Authors: | Wang, Z, Liu, J, Walker, N. | | Deposit date: | 2003-03-26 | | Release date: | 2003-06-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Function of Nurr1 identifies a Class of Ligand-Independent Nuclear Receptors
Nature, 423, 2003
|
|
6EHT
 
 | | Modulation of PCNA sliding surface by p15PAF suggests a suppressive mechanism for cisplatin-induced DNA lesion bypass by pol eta holoenzyme | | Descriptor: | DNA (5'-D(P*AP*TP*AP*CP*GP*AP*TP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*AP*TP*CP*GP*TP*AP*T)-3'), PCNA-associated factor, ... | | Authors: | De March, M, Barrera-Vilarmau, S, Mentegari, E, Merino, N, Bressan, E, Maga, G, Crehuet, R, Onesti, S, Blanco, F.J, De Biasio, A. | | Deposit date: | 2017-09-15 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | p15PAF binding to PCNA modulates the DNA sliding surface.
Nucleic Acids Res., 46, 2018
|
|
6N9V
 
 | | Structure of bacteriophage T7 lagging-strand DNA polymerase (D5A/E7A) and gp4 (helicase/primase) bound to DNA including RNA/DNA hybrid, and an incoming dTTP (LagS1) | | Descriptor: | DNA primase/helicase, DNA-directed DNA polymerase, MAGNESIUM ION, ... | | Authors: | Gao, Y, Fox, T, Val, N, Yang, W. | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-06 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
1P29
 
 | | Crystal Structure of glycogen phosphorylase b in complex with maltopentaose | | Descriptor: | Glycogen phosphorylase, muscle form, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Pinotsis, N, Leonidas, D.D, Chrysina, E.D, Oikonomakos, N.G, Mavridis, I.M. | | Deposit date: | 2003-04-15 | | Release date: | 2003-09-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The binding of beta- and gamma-cyclodextrins to glycogen phosphorylase b: Kinetic and crystallographic studies.
Protein Sci., 12, 2003
|
|
1P7N
 
 | | Dimeric Rous Sarcoma virus Capsid protein structure with an upstream 25-amino acid residue extension of C-terminal of Gag p10 protein | | Descriptor: | GAG POLYPROTEIN CAPSID PROTEIN P27 | | Authors: | Nandhagopal, N, Simpson, A.A, Johnson, M.C, Francisco, A.B, Schatz, G.W, Rossmann, M.G, Vogt, V.M. | | Deposit date: | 2003-05-02 | | Release date: | 2003-12-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dimeric rous sarcoma virus capsid protein structure relevant to immature gag assembly
J.Mol.Biol., 335, 2004
|
|
1ONL
 
 | | Crystal structure of Thermus thermophilus HB8 H-protein of the glycine cleavage system | | Descriptor: | glycine cleavage system H protein | | Authors: | Nakai, T, Ishijima, J, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-28 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Thermus thermophilus HB8 H-protein of the glycine-cleavage system, resolved by a six-dimensional molecular-replacement method.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
6N9U
 
 | | Structure of bacteriophage T7 lagging-strand DNA polymerase (D5A/E7A) interacting with primase domains of two gp4 subunits bound to an RNA/DNA hybrid and dTTP (from LagS1) | | Descriptor: | DNA (44-MER), DNA primase/helicase, DNA-directed DNA polymerase, ... | | Authors: | Gao, Y, Fox, T, Val, N, Yang, W. | | Deposit date: | 2018-12-04 | | Release date: | 2019-03-06 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structures and operating principles of the replisome.
Science, 363, 2019
|
|
1Q67
 
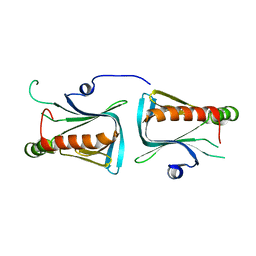 | | Crystal structure of Dcp1p | | Descriptor: | Decapping protein involved in mRNA degradation-Dcp1p | | Authors: | She, M, Decker, C.J, Liu, Y, Chen, N, Parker, R, Song, H. | | Deposit date: | 2003-08-12 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Dcp1p and its functional implications in mRNA decapping
Nat.Struct.Mol.Biol., 11, 2004
|
|
1DZE
 
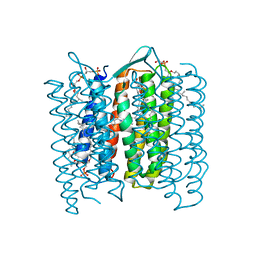 | | Structure of the M Intermediate of Bacteriorhodopsin trapped at 100K | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, 3-PHOSPHORYL-[1,2-DI-PHYTANYL]GLYCEROL, ... | | Authors: | Takeda, K, Matsui, Y, Sato, H, Hino, T, Kanamori, E, Okumura, H, Yamane, T, Iizuka, T, Kamiya, N, Adachi, S, Kouyama, T. | | Deposit date: | 2000-02-25 | | Release date: | 2000-08-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the M Intermediate of Bacteriorhodopsin: Allosteric Structural Changes Mediated by Sliding Movement of a Transmembrane Helix
J.Mol.Biol., 341, 2004
|
|
1POU
 
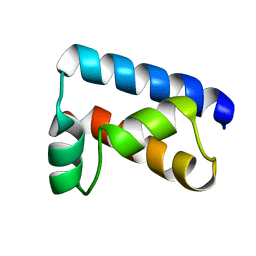 | | THE SOLUTION STRUCTURE OF THE OCT-1 POU-SPECIFIC DOMAIN REVEALS A STRIKING SIMILARITY TO THE BACTERIOPHAGE LAMBDA REPRESSOR DNA-BINDING DOMAIN | | Descriptor: | OCT-1 | | Authors: | Assa-Munt, N, Mortishire-Smith, R.J, Aurora, R, Herr, W, Wright, P.E. | | Deposit date: | 1993-06-14 | | Release date: | 1994-10-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the Oct-1 POU-specific domain reveals a striking similarity to the bacteriophage lambda repressor DNA-binding domain.
Cell(Cambridge,Mass.), 73, 1993
|
|
