1BX6
 
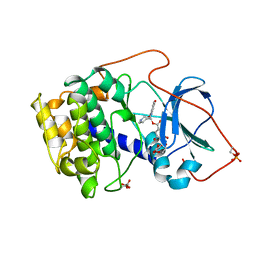 | | CRYSTAL STRUCTURE OF THE POTENT NATURAL PRODUCT INHIBITOR BALANOL IN COMPLEX WITH THE CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE | | Descriptor: | BALANOL, CAMP-DEPENDENT PROTEIN KINASE | | Authors: | Narayana, N, Xuong, N.-H, Ten Eyck, L.F, Taylor, S.S. | | Deposit date: | 1998-10-13 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the potent natural product inhibitor balanol in complex with the catalytic subunit of cAMP-dependent protein kinase.
Biochemistry, 38, 1999
|
|
1C53
 
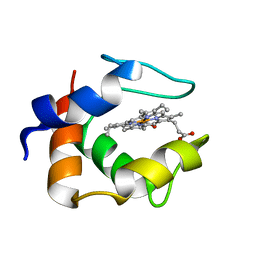 | | S-CLASS CYTOCHROMES C HAVE A VARIETY OF FOLDING PATTERNS: STRUCTURE OF CYTOCHROME C-553 FROM DESULFOVIBRIO VULGARIS DETERMINED BY THE MULTI-WAVELENGTH ANOMALOUS DISPERSION METHOD | | Descriptor: | CYTOCHROME C553, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nakagawa, A, Higuchi, Y, Yasuoka, N, Katsube, Y, Yaga, T. | | Deposit date: | 1991-08-26 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | S-class cytochromes c have a variety of folding patterns: structure of cytochrome c-553 from Desulfovibrio vulgaris determined by the multi-wavelength anomalous dispersion method.
J.Biochem.(Tokyo), 108, 1990
|
|
1APA
 
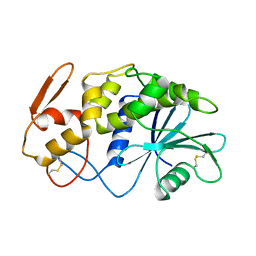 | | X-RAY STRUCTURE OF A POKEWEED ANTIVIRAL PROTEIN, CODED BY A NEW GENOMIC CLONE, AT 0.23 NM RESOLUTION. A MODEL STRUCTURE PROVIDES A SUITABLE ELECTROSTATIC FIELD FOR SUBSTRATE BINDING. | | Descriptor: | POKEWEED ANTIVIRAL PROTEIN | | Authors: | Ago, H, Kataoka, J, Tsuge, H, Habuka, N, Inagaki, E, Noma, M, Miyano, M. | | Deposit date: | 1993-09-21 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structure of a pokeweed antiviral protein, coded by a new genomic clone, at 0.23 nm resolution. A model structure provides a suitable electrostatic field for substrate binding.
Eur.J.Biochem., 225, 1994
|
|
1BVD
 
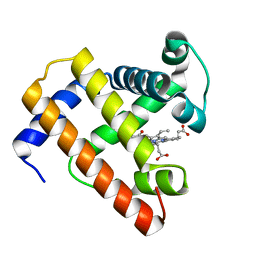 | | STRUCTURE OF A BILIVERDIN APOMYOGLOBIN COMPLEX (FORM B) AT 98 K | | Descriptor: | APOMYOGLOBIN, BILIVERDINE IX ALPHA | | Authors: | Wagner, U.G, Mueller, N, Schmitzberger, W, Falk, H, Kratky, C. | | Deposit date: | 1994-12-16 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure determination of the biliverdin apomyoglobin complex: crystal structure analysis of two crystal forms at 1.4 and 1.5 A resolution.
J.Mol.Biol., 247, 1995
|
|
1BVC
 
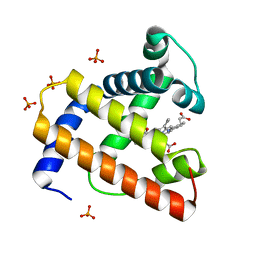 | | STRUCTURE OF A BILIVERDIN APOMYOGLOBIN COMPLEX (FORM D) AT 118 K | | Descriptor: | APOMYOGLOBIN, BILIVERDINE IX ALPHA, PHOSPHATE ION | | Authors: | Wagner, U.G, Mueller, N, Schmitzberger, W, Falk, H, Kratky, C. | | Deposit date: | 1994-12-16 | | Release date: | 1995-07-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure determination of the biliverdin apomyoglobin complex: crystal structure analysis of two crystal forms at 1.4 and 1.5 A resolution.
J.Mol.Biol., 247, 1995
|
|
1D88
 
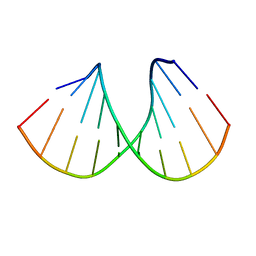 | |
1CYC
 
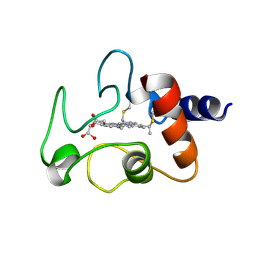 | | THE CRYSTAL STRUCTURE OF BONITO (KATSUO) FERROCYTOCHROME C AT 2.3 ANGSTROMS RESOLUTION. II. STRUCTURE AND FUNCTION | | Descriptor: | FERROCYTOCHROME C, HEME C | | Authors: | Tanaka, N, Yamane, T, Tsukihara, T, Ashida, T, Kakudo, M. | | Deposit date: | 1976-08-01 | | Release date: | 1976-10-06 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of bonito (katsuo) ferrocytochrome c at 2.3 A resolution. II. Structure and function.
J.Biochem.(Tokyo), 77, 1975
|
|
6WLC
 
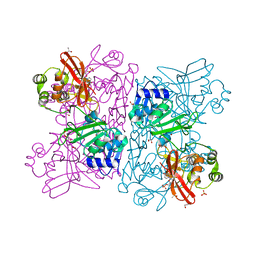 | | Crystal Structure of NSP15 Endoribonuclease from SARS CoV-2 in the Complex with Uridine-5'-Monophosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Kim, Y, Maltseva, N, Jedrzejczak, R, Endres, M, Chang, C, Godzik, A, Michalska, K, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-04-19 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Tipiracil binds to uridine site and inhibits Nsp15 endoribonuclease NendoU from SARS-CoV-2.
Commun Biol, 4, 2021
|
|
6WX9
 
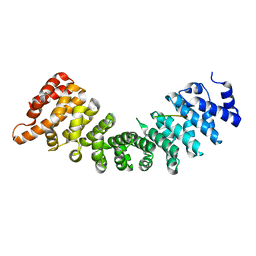 | | SOX2 bound to Importin-alpha 5 | | Descriptor: | Importin subunit alpha-5, Transcription factor SOX-2 | | Authors: | Bikshapathi, J, Stewart, M, Forwood, J.K, Aragao, D, Roman, N. | | Deposit date: | 2020-05-10 | | Release date: | 2020-10-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for nuclear import selectivity of pioneer transcription factor SOX2.
Nat Commun, 12, 2021
|
|
6ZOO
 
 | | Photosystem I reduced Plastocyanin Complex | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Nelson, N, Caspy, I, Shkolnisky, Y. | | Deposit date: | 2020-07-07 | | Release date: | 2021-06-16 | | Last modified: | 2021-07-07 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Structure of plant photosystem I-plastocyanin complex reveals strong hydrophobic interactions.
Biochem.J., 478, 2021
|
|
7PAF
 
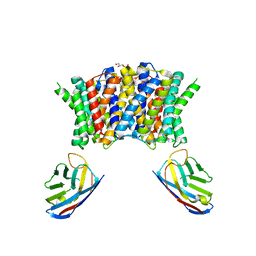 | | Streptococcus pneumoniae choline importer LicB in lipid nanodiscs | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, LicB protein, Nanobody | | Authors: | Perez, C, Baerland, N. | | Deposit date: | 2021-07-29 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Mechanistic basis of choline import involved in teichoic acids and lipopolysaccharide modification.
Sci Adv, 8, 2022
|
|
1T4O
 
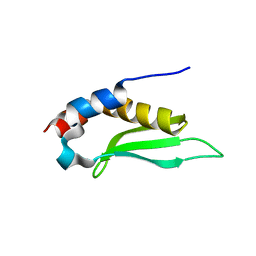 | | Crystal structure of rnt1p dsRBD | | Descriptor: | Ribonuclease III | | Authors: | Leulliot, N, Quevillon-Cheruel, S, Graille, M, van Tilbeurgh, H, Leeper, T.C, Godin, K.S, Edwards, T.E, Sigurdsson, S.T, Rozenkrants, N, Nagel, R.J, Ares Jr, M, Varani, G. | | Deposit date: | 2004-04-30 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A new alpha-helical extension promotes RNA binding by the dsRBD of Rnt1p RNAse III
Embo J., 23, 2004
|
|
1T4N
 
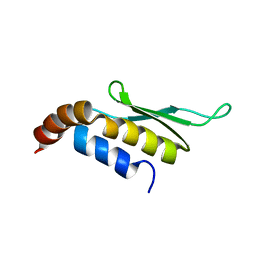 | | Solution structure of Rnt1p dsRBD | | Descriptor: | Ribonuclease III | | Authors: | Leulliot, N, Quevillon-Cheruel, S, Graille, M, van Tilbeurgh, H, Leeper, T.C, Godin, K.S, Edwards, T.E, Sigurdsson, S.T, Rozenkrants, N, Nagel, R.J, Ares, M, Varani, G. | | Deposit date: | 2004-04-30 | | Release date: | 2004-07-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A new alpha-helical extension promotes RNA binding by the dsRBD of Rnt1p RNAse III
Embo J., 23, 2004
|
|
8U7Z
 
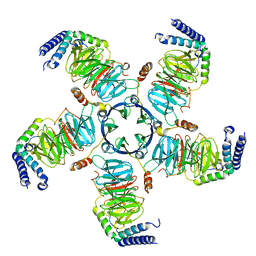 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: Local Refinment of KCTD5(CTD)/Gbeta1gamma2 | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U81
 
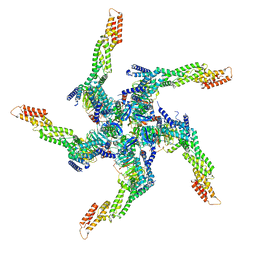 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State A From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U82
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State B From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U83
 
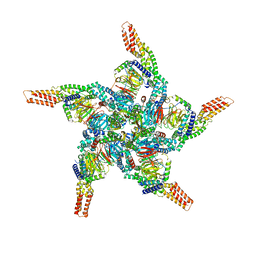 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State C From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.975 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U80
 
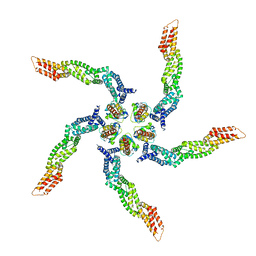 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: Local Refinment of KCTD5(BTB)/Cullin3(NTD) | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3 | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8U84
 
 | | KCTD5/Cullin3/Gbeta1gamma2 Complex: State D From Composite RELION Multi-body Refinement Map | | Descriptor: | BTB/POZ domain-containing protein KCTD5, Cullin-3, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Kuntz, D.A, Nguyen, D.M, Narayanan, N, Prive, G.G. | | Deposit date: | 2023-09-15 | | Release date: | 2023-10-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Structure and dynamics of a pentameric KCTD5/CUL3/G beta gamma E3 ubiquitin ligase complex.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6XY3
 
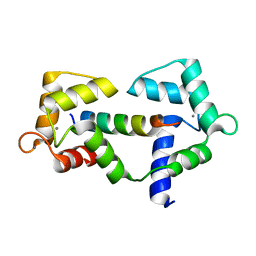 | |
6XXX
 
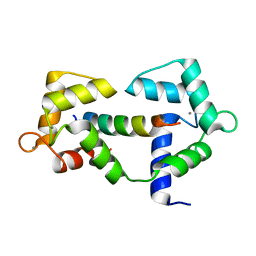 | | 1.25 Angstrom crystal structure of Ca/CaM A102V:RyR2 peptide complex | | Descriptor: | CALCIUM ION, Calmodulin-1, LYS-LYS-ALA-VAL-TRP-HIS-LYS-LEU-LEU-SER-LYS-GLN-ARG-LYS-ARG-ALA-VAL-VAL-ALA-CYS-PHE | | Authors: | Antonyuk, S, Helassa, N. | | Deposit date: | 2020-01-28 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | CPVT-associated calmodulin variants N53I and A102V dysregulate Ca2+ signalling via different mechanisms.
J.Cell.Sci., 135, 2022
|
|
6XXF
 
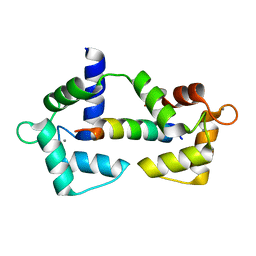 | |
1UBH
 
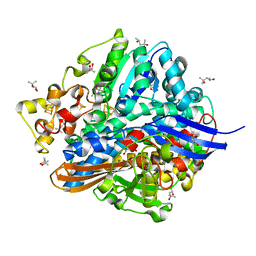 | | Three-dimensional Structure of The Carbon Monoxide Complex of [NiFe]hydrogenase From Desulufovibrio vulgaris Miyazaki F | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (MU-SULPHIDO)-BIS(MU-CYS,S)-[TRICARBONYLIRON-DI-(CYS,S)NICKEL(II)](FE-NI), CARBON MONOXIDE, ... | | Authors: | Ogata, H, Mizoguchi, Y, Mizuno, N, Miki, K, Adachi, S, Yasuoka, N, Yagi, T, Yamauchi, O, Hirota, S, Higuchi, Y. | | Deposit date: | 2003-04-04 | | Release date: | 2003-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Studies of the Carbon Monoxide Complex of [NiFe]hydrogenase from Desulfovibrio vulgaris Miyazaki F: Suggestion for the Initial Activation Site for Dihydrogen
J.Am.Chem.Soc., 124, 2002
|
|
7VLD
 
 | | Oxy-deoxy intermediate of V2 hemoglobin at 69% oxygen saturation | | Descriptor: | CALCIUM ION, Extracellular A1 globin, Extracellular A2 globin, ... | | Authors: | Numoto, N, Onoda, S, Kawano, Y, Okumura, H, Baba, S, Fukumori, Y, Miki, K, Ito, N. | | Deposit date: | 2021-10-02 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of oxygen dissociation intermediates of 400 kDa V2 hemoglobin provide coarse snapshots of the protein allostery.
Biophys Physicobio., 19, 2022
|
|
7VLF
 
 | | Oxy-deoxy intermediate of V2 hemoglobin at 26% oxygen saturation | | Descriptor: | CALCIUM ION, Extracellular A1 globin, Extracellular A2 globin, ... | | Authors: | Numoto, N, Onoda, S, Kawano, Y, Okumura, H, Baba, S, Fukumori, Y, Miki, K, Ito, N. | | Deposit date: | 2021-10-02 | | Release date: | 2022-05-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of oxygen dissociation intermediates of 400 kDa V2 hemoglobin provide coarse snapshots of the protein allostery.
Biophys Physicobio., 19, 2022
|
|
