1DZE
 
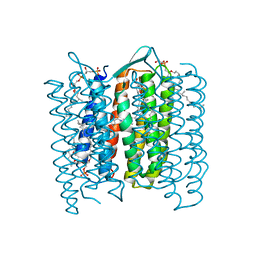 | | Structure of the M Intermediate of Bacteriorhodopsin trapped at 100K | | Descriptor: | 2,3-DI-O-PHYTANLY-3-SN-GLYCERO-1-PHOSPHORYL-3'-SN-GLYCEROL-1'-PHOSPHATE, 2,3-DI-PHYTANYL-GLYCEROL, 3-PHOSPHORYL-[1,2-DI-PHYTANYL]GLYCEROL, ... | | Authors: | Takeda, K, Matsui, Y, Sato, H, Hino, T, Kanamori, E, Okumura, H, Yamane, T, Iizuka, T, Kamiya, N, Adachi, S, Kouyama, T. | | Deposit date: | 2000-02-25 | | Release date: | 2000-08-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the M Intermediate of Bacteriorhodopsin: Allosteric Structural Changes Mediated by Sliding Movement of a Transmembrane Helix
J.Mol.Biol., 341, 2004
|
|
1UBC
 
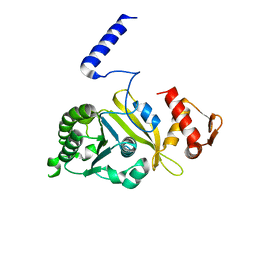 | | Structure of Reca Protein | | Descriptor: | RecA | | Authors: | Datta, S, Krishna, R, Ganesh, N, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2003-04-04 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Crystal Structures of Mycobacterium smegmatis RecA and Its Nucleotide Complexes
J.BACTERIOL., 185, 2003
|
|
1KM0
 
 | |
5XJA
 
 | | The Crystal Structure of the Minimal Core Domain of the Microtubule Depolymerizer KIF2C Complexed with ADP-Mg-AlFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Kinesin-like protein KIF2C, ... | | Authors: | Ogawa, T, Jiang, X, Hirokawa, N. | | Deposit date: | 2017-04-30 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | Mechanism of Catalytic Microtubule Depolymerization via KIF2-Tubulin Transitional Conformation
Cell Rep, 20, 2017
|
|
1KM1
 
 | |
7OGP
 
 | | Structure of the apo-state of the bacteriophage PhiKZ non-virion RNA polymerase - class including clamp | | Descriptor: | DNA-directed RNA polymerase, PHIKZ055, PHIKZ068, ... | | Authors: | de Martin Garrido, N, Lai Wan Loong, Y.T.E, Yakunina, M, Aylett, C.H.S. | | Deposit date: | 2021-05-07 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the bacteriophage PhiKZ non-virion RNA polymerase.
Nucleic Acids Res., 49, 2021
|
|
7OGR
 
 | | Structure of the apo-state of the bacteriophage PhiKZ non-virion RNA polymerase | | Descriptor: | DNA-directed RNA polymerase, PHIKZ055, PHIKZ068, ... | | Authors: | de Martin Garrido, N, Lai Wan Loong, Y.T.E, Yakunina, M, Aylett, C.H.S. | | Deposit date: | 2021-05-07 | | Release date: | 2021-07-07 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the bacteriophage PhiKZ non-virion RNA polymerase.
Nucleic Acids Res., 49, 2021
|
|
1Y5I
 
 | | The crystal structure of the NarGHI mutant NarI-K86A | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, FE3-S4 CLUSTER, ... | | Authors: | Bertero, M.G, Rothery, R.A, Boroumand, N, Palak, M, Blasco, F, Ginet, N, Weiner, J.H, Strynadka, N.C.J. | | Deposit date: | 2004-12-02 | | Release date: | 2005-03-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Biochemical Characterization of a Quinol Binding Site of Escherichia coli Nitrate Reductase A
J.Biol.Chem., 280, 2005
|
|
6HE2
 
 | | Crystal structure of an open conformation of 2-Hydroxyisobutyryl-CoA Ligase (HCL) in complex with 2-HIB-AMP and CoA | | Descriptor: | 2-hydroxyisobutyryl-CoA synthetase, ADENOSINE MONOPHOSPHATE, COENZYME A, ... | | Authors: | Zahn, M, Rohwerder, T, Strater, N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of 2-Hydroxyisobutyric Acid-CoA Ligase Reveal Determinants of Substrate Specificity and Describe a Multi-Conformational Catalytic Cycle.
J.Mol.Biol., 431, 2019
|
|
1KLY
 
 | |
1KM4
 
 | |
1F9A
 
 | | CRYSTAL STRUCTURE ANALYSIS OF NMN ADENYLYLTRANSFERASE FROM METHANOCOCCUS JANNASCHII | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, HYPOTHETICAL PROTEIN MJ0541, MAGNESIUM ION | | Authors: | D'Angelo, I, Raffaelli, N, Dabusti, V, Lorenzi, T, Magni, G, Rizzi, M. | | Deposit date: | 2000-07-09 | | Release date: | 2001-01-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of nicotinamide mononucleotide adenylyltransferase: a key enzyme in NAD(+) biosynthesis.
Structure Fold.Des., 8, 2000
|
|
1SKM
 
 | | HhaI methyltransferase in complex with DNA containing an abasic south carbocyclic sugar at its target site | | Descriptor: | 5'-D(*T*GP*TP*CP*AP*GP*(HCX)P*GP*CP*AP*TP*GP*G)-3', 5'-D(*TP*CP*CP*AP*TP*GP*CP*GP*CP*TP*GP*AP*C)-3', Modification methylase HhaI, ... | | Authors: | Horton, J.R, Ratner, G, Banavali, N, Huang, N, Marquez, V.E, MacKerell, A.D, Cheng, X. | | Deposit date: | 2004-03-05 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Caught in the act: visualization of an intermediate in the DNA base-flipping pathway induced by HhaI methyltransferase
Nucleic Acids Res., 32, 2004
|
|
6AEJ
 
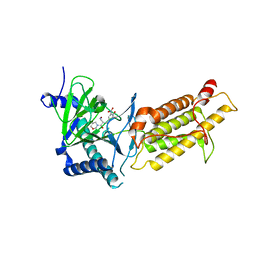 | | Crystal structure of human FTO in complex with small-molecule inhibitors | | Descriptor: | (E)-3-[3-nitro-4,5-bis(oxidanyl)phenyl]-2-(1,3-oxazinan-3-ylcarbonyl)prop-2-enenitrile, Alpha-ketoglutarate-dependent dioxygenase FTO,Alpha-ketoglutarate-dependent dioxygenase FTO, ZINC ION | | Authors: | Wang, Y, Cao, R, Peng, S, Zhang, W, Huang, N. | | Deposit date: | 2018-08-05 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1.
Sci Transl Med, 11, 2019
|
|
1KKD
 
 | | Solution structure of the calmodulin binding domain (CaMBD) of small conductance Ca2+-activated potassium channels (SK2) | | Descriptor: | Small conductance calcium-activated potassium channel protein 2 | | Authors: | Wissmann, R, Bildl, W, Neumann, H, Rivard, A.F, Kloecker, N, Weitz, D, Schulte, U, Adelman, J.P, Bentrop, D, Fakler, B. | | Deposit date: | 2001-12-07 | | Release date: | 2001-12-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A helical region in the C terminus of small-conductance Ca2+-activated K+ channels controls assembly with apo-calmodulin.
J.Biol.Chem., 277, 2002
|
|
1KC8
 
 | | Co-crystal Structure of Blasticidin S Bound to the 50S Ribosomal Subunit | | Descriptor: | 23S RRNA, 5S RRNA, BLASTICIDIN S, ... | | Authors: | Hansen, J.L, Ban, N, Nissen, P, Moore, P.B, Steitz, T.A. | | Deposit date: | 2001-11-07 | | Release date: | 2003-07-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structures of Five Antibiotics Bound at the Peptidyl Transferase Center of
the Large Ribosomal Subunit
J.Mol.Biol., 330, 2003
|
|
1FFS
 
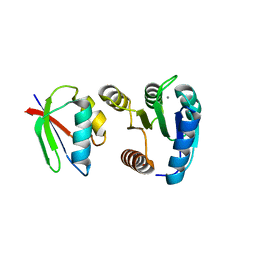 | | CHEY-BINDING DOMAIN OF CHEA IN COMPLEX WITH CHEY FROM CRYSTALS SOAKED IN ACETYL PHOSPHATE | | Descriptor: | CHEMOTAXIS PROTEIN CHEA, CHEMOTAXIS PROTEIN CHEY, MANGANESE (II) ION | | Authors: | Gouet, P, Chinardet, N, Welch, M, Guillet, V, Birck, C, Mourey, L, Samama, J.-P. | | Deposit date: | 2000-07-26 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Further insights into the mechanism of function of the response regulator CheY from crystallographic studies of the CheY--CheA(124--257) complex.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1Z7R
 
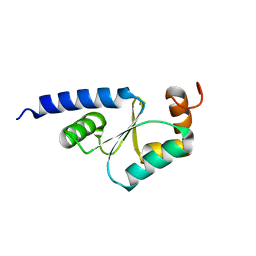 | | Solution Structure of reduced glutaredoxin C1 from Populus tremula x tremuloides | | Descriptor: | glutaredoxin | | Authors: | Feng, Y, Zhong, N, Rouhier, N, Jacquot, J.P, Xia, B. | | Deposit date: | 2005-03-26 | | Release date: | 2006-03-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural Insight into Poplar Glutaredoxin C1 with a Bridging Iron-Sulfur Cluster at the Active Site
Biochemistry, 45, 2006
|
|
7OBR
 
 | | RNC-SRP early complex | | Descriptor: | 28S rRNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Jomaa, A, Ban, N. | | Deposit date: | 2021-04-23 | | Release date: | 2021-07-21 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular mechanism of cargo recognition and handover by the mammalian signal recognition particle.
Cell Rep, 36, 2021
|
|
7OBQ
 
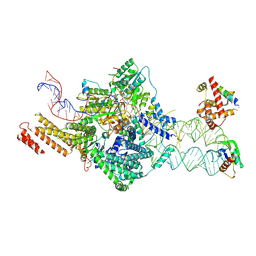 | | SRP-SR at the distal site conformation | | Descriptor: | EM14S01-3B_G0054400.mRNA.1.CDS.1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Jomaa, A, Ban, N. | | Deposit date: | 2021-04-23 | | Release date: | 2021-07-21 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Molecular mechanism of cargo recognition and handover by the mammalian signal recognition particle.
Cell Rep, 36, 2021
|
|
1FPN
 
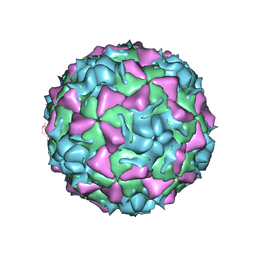 | | HUMAN RHINOVIRUS SEROTYPE 2 (HRV2) | | Descriptor: | COAT PROTEIN VP1, COAT PROTEIN VP2, COAT PROTEIN VP3, ... | | Authors: | Verdaguer, N, Blaas, D, Fita, I. | | Deposit date: | 2000-08-31 | | Release date: | 2000-09-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of human rhinovirus serotype 2 (HRV2).
J.Mol.Biol., 300, 2000
|
|
1KWG
 
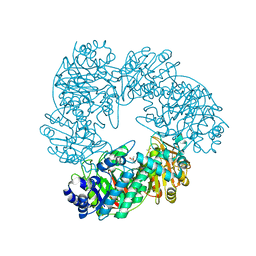 | | Crystal structure of Thermus thermophilus A4 beta-galactosidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, BETA-GALACTOSIDASE, ... | | Authors: | Hidaka, M, Fushinobu, S, Ohtsu, N, Motoshima, H, Matsuzawa, H, Shoun, H, Wakagi, T. | | Deposit date: | 2002-01-29 | | Release date: | 2002-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Trimeric Crystal Structure of the Glycoside Hydrolase Family 42 beta-Galactosidase from Thermus thermophilus A4 and the Structure of its Complex with Galactose
J.MOL.BIOL., 322, 2002
|
|
1KD1
 
 | | Co-crystal Structure of Spiramycin bound to the 50S Ribosomal Subunit of Haloarcula marismortui | | Descriptor: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | Authors: | Hansen, J.L, Ippolito, J.A, Ban, N, Nissen, P, Moore, P.B, Steitz, T.A. | | Deposit date: | 2001-11-12 | | Release date: | 2002-07-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structures of four macrolide antibiotics bound to the large ribosomal subunit.
Mol.Cell, 10, 2002
|
|
1KVD
 
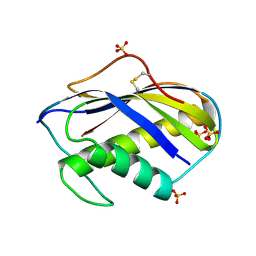 | | KILLER TOXIN FROM HALOTOLERANT YEAST | | Descriptor: | SMK TOXIN, SULFATE ION | | Authors: | Kashiwagi, T, Kunishima, N, Suzuki, C, Tsuchiya, F, Nikkuni, S, Arata, Y, Morikawa, K. | | Deposit date: | 1996-10-04 | | Release date: | 1997-04-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The novel acidophilic structure of the killer toxin from halotolerant yeast demonstrates remarkable folding similarity with a fungal killer toxin.
Structure, 5, 1997
|
|
1KM2
 
 | |
