8BEF
 
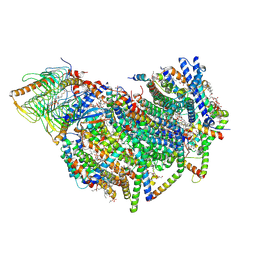 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CI membrane core) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.13 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
8BEP
 
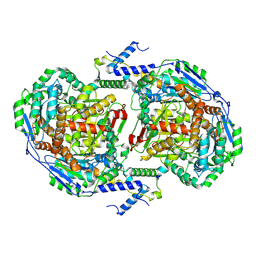 | |
1TAM
 
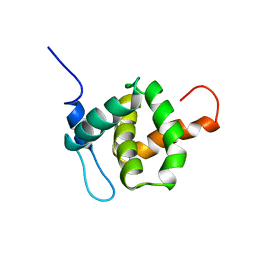 | | HUMAN IMMUNODEFICIENCY VIRUS, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | HIV-1 MATRIX PROTEIN | | Authors: | Matthews, S, Barlow, P, Clark, N, Kingsman, S, Kingsman, A, Campbell, I. | | Deposit date: | 1996-02-07 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of p17, the HIV matrix protein.
Biochem.Soc.Trans., 23, 1995
|
|
8BED
 
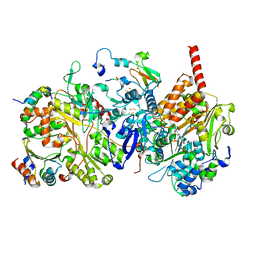 | |
8BEE
 
 | |
8BPX
 
 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Complete composition) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-11-18 | | Release date: | 2023-01-25 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.09 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
4TNI
 
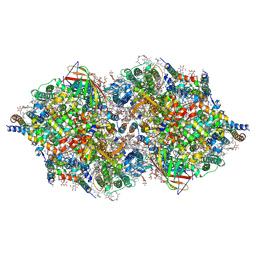 | | RT XFEL structure of Photosystem II 500 ms after the third illumination at 4.6 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (4.6 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
1IUZ
 
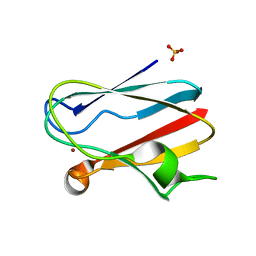 | | PLASTOCYANIN | | Descriptor: | COPPER (II) ION, PLASTOCYANIN, SULFATE ION | | Authors: | Shibata, N. | | Deposit date: | 1996-10-06 | | Release date: | 1997-08-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Novel insight into the copper-ligand geometry in the crystal structure of Ulva pertusa plastocyanin at 1.6-A resolution. Structural basis for regulation of the copper site by residue 88.
J.Biol.Chem., 274, 1999
|
|
8BVG
 
 | |
5U94
 
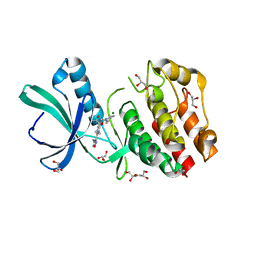 | | Crystal structure of the Mycobacterium tuberculosis PASTA kinase PknB in complex with the potential theraputic kinase inhibitor GSK690693. | | Descriptor: | 4-{2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-[(3S)-piperidin-3-ylmethoxy]-1H-imidazo[4,5-c]pyridin-4-yl}-2-methylbut-3 -yn-2-ol, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Wlodarchak, N, Satyshur, K, Striker, R. | | Deposit date: | 2016-12-15 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | In Silico Screen and Structural Analysis Identifies Bacterial Kinase Inhibitors which Act with beta-Lactams To Inhibit Mycobacterial Growth.
Mol. Pharm., 15, 2018
|
|
4TNK
 
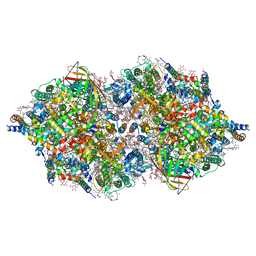 | | RT XFEL structure of Photosystem II 250 microsec after the third illumination at 5.2 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (5.2 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
4THN
 
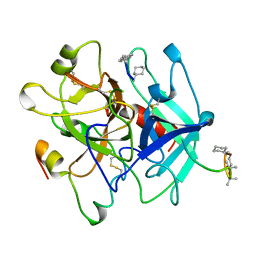 | | THE CRYSTAL STRUCTURE OF ALPHA-THROMBIN-HIRUNORM IV COMPLEX REVEALS A NOVEL SPECIFICITY SITE RECOGNITION MODE. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-THROMBIN, HIRUNORM IV | | Authors: | Lombardi, A, De Simone, G, Nastri, F, Galdiero, S, Della Morte, R, Staiano, N, Pedone, C, Bolognesi, M, Pavone, V. | | Deposit date: | 1998-09-18 | | Release date: | 1999-06-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of alpha-thrombin-hirunorm IV complex reveals a novel specificity site recognition mode.
Protein Sci., 8, 1999
|
|
1UAT
 
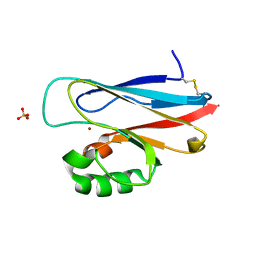 | | The significance of the flexible loop in the azurin (Az-iso2) from the obligate methylotroph Methylomonas sp. strain J | | Descriptor: | Azurin iso-2, COPPER (II) ION, SULFATE ION | | Authors: | Inoue, T, Suzuki, S, Nisho, N, Yamaguchi, K, Kataoka, K, Tobari, J, Yong, X, Hamanaka, S, Matsumura, H, Kai, Y. | | Deposit date: | 2003-03-19 | | Release date: | 2004-03-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The significance of the flexible loop in the azurin (Az-iso2) from the obligate methylotroph Methylomonas sp. strain J
J.Mol.Biol., 333, 2003
|
|
8BQ6
 
 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (Complete conformation 2 composition) | | Descriptor: | 2,3-DIMETHOXY-5-METHYL-6-(3,11,15,19-TETRAMETHYL-EICOSA-2,6,10,14,18-PENTAENYL)-[1,4]BENZOQUINONE, AT3G07480.1, Acyl carrier protein 1, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-11-18 | | Release date: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
5CS2
 
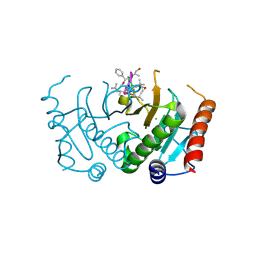 | | Crystal structure of Plasmodium falciparum diadenosine triphosphate hydrolase in complex with Cyclomarin A | | Descriptor: | CHLORIDE ION, Cyclomarin A, Histidine triad protein | | Authors: | Ostermann, N, Schmitt, E, Gerhartz, B, Hinniger, A, Delmas, C. | | Deposit date: | 2015-07-23 | | Release date: | 2015-10-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Gift from Nature: Cyclomarin A Kills Mycobacteria and Malaria Parasites by Distinct Modes of Action.
Chembiochem, 16, 2015
|
|
7MYZ
 
 | |
1P4C
 
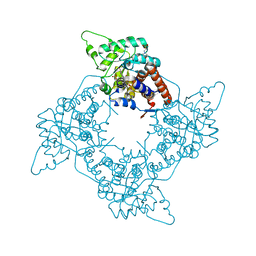 | | High Resolution Structure of Oxidized Active Mutant of (S)-Mandelate Dehydrogenase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN MONONUCLEOTIDE, L(+)-Mandelate Dehydrogenase, ... | | Authors: | Sukumar, N, Mitra, B, Mathews, F.S. | | Deposit date: | 2003-04-22 | | Release date: | 2003-10-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | High Resolution Structures of an Oxidized and Reduced Flavoprotein: THE WATER SWITCH IN A SOLUBLE FORM OF (S)-MANDELATE DEHYDROGENASE
J.Biol.Chem., 279, 2004
|
|
6FQB
 
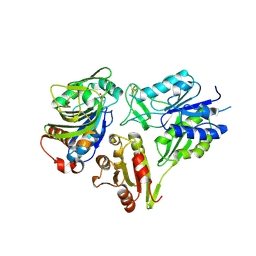 | | MurT/GatD peptidoglycan amidotransferase complex from Streptococcus pneumoniae R6 | | Descriptor: | Cobyric acid synthase, GLUTAMINE, Mur ligase family protein | | Authors: | Morlot, C, Contreras-Martel, C, Leisico, F, Straume, D, Peters, K, Hegnar, O.A, Simon, N, Villard, A.M, Breukink, E, Gravier-Pelletier, C, Le Corre, L, Vollmer, W, Pietrancosta, N, Havarstein, L.S, Zapun, A. | | Deposit date: | 2018-02-13 | | Release date: | 2018-08-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the essential peptidoglycan amidotransferase MurT/GatD complex from Streptococcus pneumoniae.
Nat Commun, 9, 2018
|
|
4ZAR
 
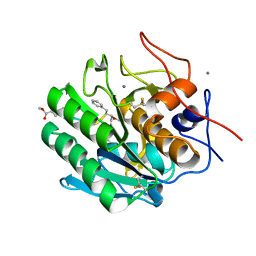 | | Crystal Structure of Proteinase K from Engyodontium albuminhibited by METHOXYSUCCINYL-ALA-ALA-PRO-PHE-CHLOROMETHYL KETONE at 1.15 A resolution | | Descriptor: | CALCIUM ION, METHOXYSUCCINYL-ALA-ALA-PRO-PHE-CHLOROMETHYL KETONE, bound form, ... | | Authors: | Sawaya, M.R, Cascio, D, Collazo, M, Bond, C, Cohen, A, DeNicola, A, Eden, K, Jain, K, Leung, C, Lubock, N, McCormick, J, Rosinski, J, Spiegelman, L, Athar, Y, Tibrewal, N, Winter, J, Solomon, S. | | Deposit date: | 2015-04-14 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structure of Proteinase K from Engyodontium album inhibited by METHOXYSUCCINYL-ALA-ALA-PRO-PHE-CHLOROMETHYL KETONE at 1.15 A resolution
to be published
|
|
2RTU
 
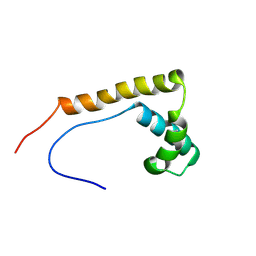 | | Solution structure of oxidized human HMGB1 A box | | Descriptor: | High mobility group protein B1 | | Authors: | Jing, W, Tochio, N, Tate, S. | | Deposit date: | 2013-09-12 | | Release date: | 2014-03-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Redox-sensitive structural change in the A-domain of HMGB1 and its implication for the binding to cisplatin modified DNA.
Biochem.Biophys.Res.Commun., 441, 2013
|
|
8B9S
 
 | |
7VDN
 
 | | High resolution crystal structure of Sperm Whale Myoglobin in the carbonmonoxy form | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Shibayama, N, Sato-Tomita, A, Ishimoto, N, Park, S.Y. | | Deposit date: | 2021-09-07 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (0.93 Å) | | Cite: | X-ray fluorescence holography of biological metal sites: Application to myoglobin.
Biochem.Biophys.Res.Commun., 635, 2022
|
|
4X35
 
 | | A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Roedig, P, Vartiainen, I, Duman, R, Panneerselvam, S, Stuebe, N, Lorbeer, O, Warmer, M, Sutton, G, Stuart, D.I, Weckert, E, David, C, Wagner, A, Meents, A. | | Deposit date: | 2014-11-27 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering.
Sci Rep, 5, 2015
|
|
1J2W
 
 | | Tetrameric Structure of aldolase from Thermus thermophilus HB8 | | Descriptor: | Aldolase protein | | Authors: | Lokanath, N.K, Shiromizu, I, Miyano, M, Yokoyama, S, Kuramitsu, S, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-01-14 | | Release date: | 2003-04-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of aldolase from Thermus thermophilus HB8 showing the contribution of oligomeric state to thermostability.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1DPR
 
 | | STRUCTURES OF THE APO-AND METAL ION ACTIVATED FORMS OF THE DIPHTHERIA TOX REPRESSOR FROM CORYNEBACTERIUM DIPHTHERIAE | | Descriptor: | DIPHTHERIA TOX REPRESSOR | | Authors: | Schiering, N, Tao, X, Murphy, J, Petsko, G.A, Ringe, D. | | Deposit date: | 1995-02-06 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the apo- and the metal ion-activated forms of the diphtheria tox repressor from Corynebacterium diphtheriae.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
