8WPK
 
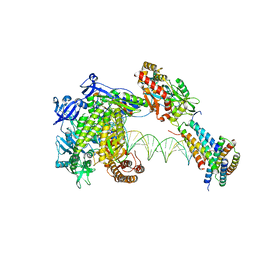 | | Structure of monkeypox virus polymerase complex F8-A22-E4-H5 with exgenous DNA | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, DNA polymerase, DNA polymerase processivity factor, ... | | Authors: | Wang, X, Li, N, Gao, N. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into the assembly and mechanism of mpox virus DNA polymerase complex F8-A22-E4-H5.
Mol.Cell, 83, 2023
|
|
8WPP
 
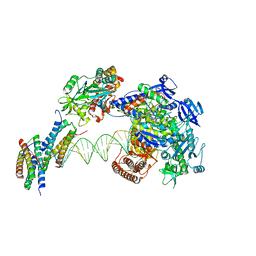 | | Structure of monkeypox virus polymerase complex F8-A22-E4-H5 with endogenous DNA | | Descriptor: | A22R DNA polymerase processivity factor, DNA polymerase, E4R Uracil-DNA glycosylase, ... | | Authors: | Wang, X, Li, N, Gao, N. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the assembly and mechanism of mpox virus DNA polymerase complex F8-A22-E4-H5.
Mol.Cell, 83, 2023
|
|
8XGC
 
 | | Structure of yeast replisome associated with FACT and histone hexamer, Composite map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, Chromosome segregation in meiosis protein 3, ... | | Authors: | Li, N, Gao, Y, Yu, D, Gao, N, Zhai, Y. | | Deposit date: | 2023-12-15 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Parental histone transfer caught at the replication fork.
Nature, 627, 2024
|
|
7VQP
 
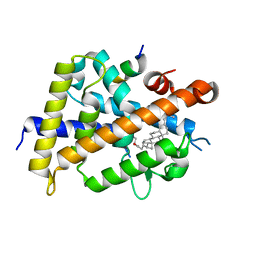 | | Vitamin D receptor complexed with a lithocholic acid derivative | | Descriptor: | 3-((R)-4-((3R,5R,8R,9S,10S,13R,14S,17R)-3-(2-hydroxy-2-methylpropyl)-10,13-dimethylhexadecahydro-1H-cyclopenta[a]phenanthren-17-yl)pentanamido)propanoic acid, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Kato, K, Numoto, N, Kagechika, H, Tanatani, A, Ito, N. | | Deposit date: | 2021-10-20 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Lithocholic Acid Amides as Potent Vitamin D Receptor Agonists.
Biomolecules, 12, 2022
|
|
7BPM
 
 | | Solution NMR structure of NF2; de novo designed protein with a novel fold | | Descriptor: | NF2 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
7BQD
 
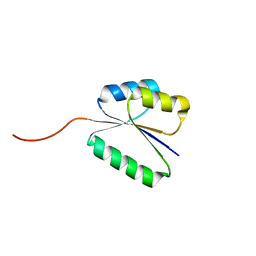 | | Solution NMR structure of NF8 (knot fold); de novo designed protein with a novel fold | | Descriptor: | NF8 | | Authors: | Kobayashi, N, Sugiki, T, Fujiwara, T, Minami, S, Koga, R, Chikenji, G, Koga, N. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Exploration of novel alpha-beta-protein folds through de novo design
Nat.Struct.Mol.Biol., 2023
|
|
8UDU
 
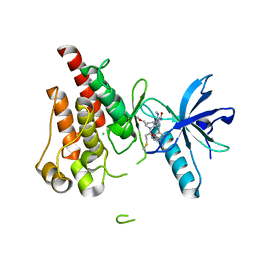 | | The X-RAY co-crystal structure of human FGFR3 and Compound 17 | | Descriptor: | 3-[(6-chloro-1-cyclopropyl-1H-benzimidazol-5-yl)ethynyl]-1-[(3S,5S)-5-(methoxymethyl)-1-(prop-2-enoyl)pyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, CHLORIDE ION, Fibroblast growth factor receptor 3 | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.737 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
1JWF
 
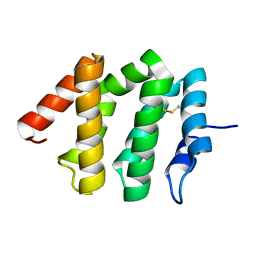 | | Crystal Structure of human GGA1 VHS domain. | | Descriptor: | ADP-ribosylation factor binding protein GGA1 | | Authors: | Shiba, T, Takatsu, H, Nogi, T, Matsugaki, N, Kawasaki, M, Igarashi, N, Suzuki, M, Kato, R, Earnest, T, Nakayama, K, Wakatsuki, S. | | Deposit date: | 2001-09-04 | | Release date: | 2002-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for recognition of acidic-cluster dileucine sequence by GGA1.
Nature, 415, 2002
|
|
8UDT
 
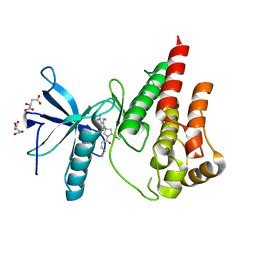 | | The X-RAY co-crystal structure of human FGFR3 and KIN-3248 | | Descriptor: | 3-[(1-cyclopropyl-4,6-difluoro-1H-benzimidazol-5-yl)ethynyl]-1-[(3R,5R)-5-(methoxymethyl)-1-propanoylpyrrolidin-3-yl]-5-(methylamino)-1H-pyrazole-4-carboxamide, D-MALATE, Fibroblast growth factor receptor 3 | | Authors: | Tyhonas, J.S, Arnold, L.D, Cox, J, Franovic, A, Gardiner, E, Grandinetti, K, Kania, R, Kanouni, T, Lardy, M, Li, C, Martin, E.S, Miller, N, Mohan, A, Murphy, E.A, Perez, M, Soroceanu, L, Timple, N, Uryu, S, Womble, S, Kaldor, S.W. | | Deposit date: | 2023-09-29 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.829 Å) | | Cite: | Discovery of KIN-3248, An Irreversible, Next Generation FGFR Inhibitor for the Treatment of Advanced Tumors Harboring FGFR2 and/or FGFR3 Gene Alterations.
J.Med.Chem., 67, 2024
|
|
8RP8
 
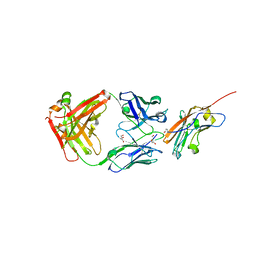 | | Structure of K2 Fab in complex with human CD47 ECD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Laursen, M, Kelpsas, V, Rose, N. | | Deposit date: | 2024-01-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural analysis of light chain-driven bispecific antibodies targeting CD47 and PD-L1.
Mabs, 16, 2024
|
|
8XGV
 
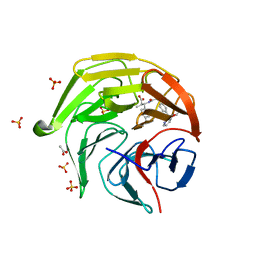 | | Optimization Efforts for Identification of Novel Highly Potent Keap1-Nrf2 Protein-Protein Interaction (PPI) Inhibitors | | Descriptor: | (2~{R},3~{S})-3-[[(2~{S})-2-[4-[(3-ethoxypyridin-2-yl)methyl]phenyl]-2-fluoranyl-ethanoyl]amino]-2-methyl-3-(4-methylphenyl)propanoic acid, ACETATE ION, Kelch-like ECH-associated protein 1, ... | | Authors: | Otake, K, Hara, Y, Ubukata, M, Inoue, M, Nagahashi, N, Motoda, D, Ogawa, N, Hantani, Y, Hantani, R, Adachi, T, Nomura, A, Yamaguchi, K, Maekawa, M, Mamada, H, Motomura, T, Sato, M, Harada, K. | | Deposit date: | 2023-12-15 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Optimization Efforts for Identification of Novel Highly Potent Keap1-Nrf2 Protein-Protein Interaction Inhibitors.
J.Med.Chem., 67, 2024
|
|
1IT7
 
 | | Crystal structure of archaeosine tRNA-guanine transglycosylase complexed with guanine | | Descriptor: | Archaeosine tRNA-guanine transglycosylase, GUANINE, MAGNESIUM ION, ... | | Authors: | Ishitani, R, Nureki, O, Fukai, S, Kijimoto, T, Nameki, N, Watanabe, M, Kondo, H, Sekine, M, Okada, N, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-01-11 | | Release date: | 2002-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of archaeosine tRNA-guanine transglycosylase.
J.Mol.Biol., 318, 2002
|
|
1IT8
 
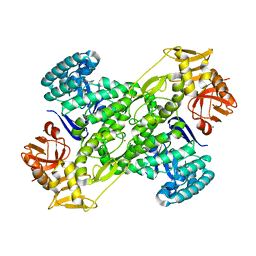 | | Crystal structure of archaeosine tRNA-guanine transglycosylase from Pyrococcus horikoshii complexed with archaeosine precursor, preQ0 | | Descriptor: | 2-AMINO-4-OXO-4,7-DIHYDRO-3H-PYRROLO[2,3-D]PYRIMIDINE-5-CARBONITRILE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Ishitani, R, Nureki, O, Fukai, S, Kijimoto, T, Nameki, N, Watanabe, M, Kondo, H, Sekine, M, Okada, N, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-01-11 | | Release date: | 2002-05-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of archaeosine tRNA-guanine transglycosylase.
J.Mol.Biol., 318, 2002
|
|
1I7Y
 
 | |
2R5W
 
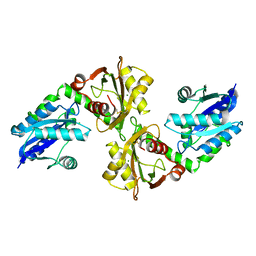 | | Crystal structure of a bifunctional NMN adenylyltransferase/ADP ribose pyrophosphatase from Francisella tularensis | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Nicotinamide-nucleotide adenylyltransferase | | Authors: | Huang, N, Sorci, L, Zhang, X, Brautigan, C, Li, X, Raffaelli, N, Grishin, N, Osterman, A, Zhang, H. | | Deposit date: | 2007-09-04 | | Release date: | 2008-03-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bifunctional NMN Adenylyltransferase/ADP-Ribose Pyrophosphatase: Structure and Function in Bacterial NAD Metabolism.
Structure, 16, 2008
|
|
1JJP
 
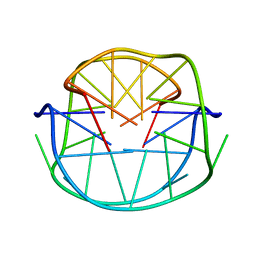 | | A(GGGG) Pentad-Containing Dimeric DNA Quadruplex Involving Stacked G(anti)G(anti)G(anti)G(syn) Tetrads | | Descriptor: | 5'-D(*GP*GP*GP*AP*GP*GP*TP*TP*TP*GP*GP*GP*AP*T)-3' | | Authors: | Zhang, N, Gorin, A, Majumdar, A, Kettani, A, Chernichenko, N, Skripkin, E, Patel, D.J. | | Deposit date: | 2001-07-09 | | Release date: | 2001-09-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | V-shaped scaffold: a new architectural motif identified in an A x (G x G x G x G) pentad-containing dimeric DNA quadruplex involving stacked G(anti) x G(anti) x G(anti) x G(syn) tetrads.
J.Mol.Biol., 311, 2001
|
|
1JVC
 
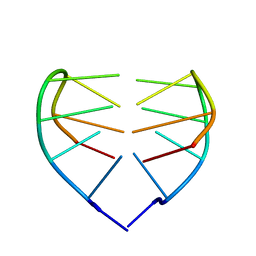 | | Dimeric DNA Quadruplex Containing Major Groove-Aligned A.T.A.T and G.C.G.C Tetrads Stabilized by Inter-Subunit Watson-Crick A:T and G:C Pairs | | Descriptor: | 5'-D(*GP*AP*GP*CP*AP*GP*GP*T)-3' | | Authors: | Zhang, N, Gorin, A, Majumdar, A, Kettani, A, Chernichenko, N, Skripkin, E, Patel, D.J. | | Deposit date: | 2001-08-29 | | Release date: | 2001-10-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Dimeric DNA quadruplex containing major groove-aligned A-T-A-T and G-C-G-C tetrads stabilized by inter-subunit Watson-Crick A-T and G-C pairs.
J.Mol.Biol., 312, 2001
|
|
2P4T
 
 | |
1LWR
 
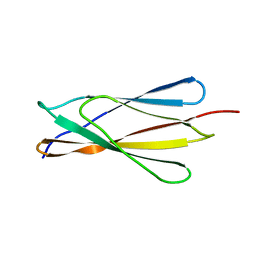 | | Solution structure of the NCAM fibronectin type III module 2 | | Descriptor: | Neural Cell Adhesion Molecule 1, 140 kDa isoform | | Authors: | Kiselyov, V.V, Skladchikova, G, Hinsby, A.M, Jensen, P.H, Kulahin, N, Pedersen, N, Tsetlin, V, Poulsen, F.M, Berezin, V, Bock, E. | | Deposit date: | 2002-06-03 | | Release date: | 2003-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for a direct interaction between FGFR1 and NCAM and evidence for a regulatory role of ATP
Structure, 11, 2003
|
|
9F1C
 
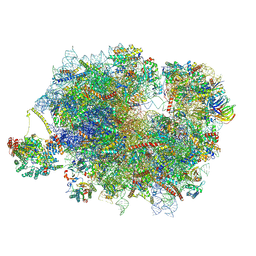 | | Mammalian quaternary complex of a translating 80S ribosome, NAC, MetAP1 and NatA/E | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S11, ... | | Authors: | Yudin, D, Scaiola, A, Ban, N. | | Deposit date: | 2024-04-18 | | Release date: | 2024-08-21 | | Last modified: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | NAC guides a ribosomal multienzyme complex for nascent protein processing.
Nature, 633, 2024
|
|
9BIY
 
 | |
9ENT
 
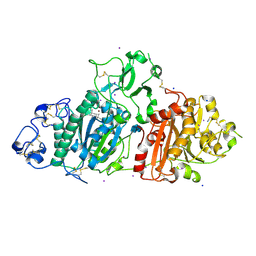 | | SSX structure of Autotaxin in cryogenic conditions | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7alpha-hydroxycholesterol, CALCIUM ION, ... | | Authors: | Eymery, M.C, McCarthy, A.A, Foos, N, Basu, S. | | Deposit date: | 2024-03-13 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | In situ serial crystallography facilitates 96-well plate structural analysis at low symmetry.
Iucrj, 11, 2024
|
|
9GDK
 
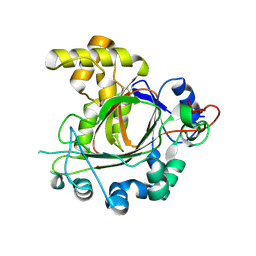 | | Jumonji domain-containing protein 1C with crystallization epitope mutations L2440Y:G2444H | | Descriptor: | Probable JmjC domain-containing histone demethylation protein 2C | | Authors: | Fairhead, M, Strain-Damerell, C, Ye, M, Mackinnon, S.R, Pinkas, D, MacLean, E.M, Koekemoer, L, Damerell, D, Krojer, T, Arrowsmith, C.H, Edwards, A, Bountra, C, Yue, W, Burgess-Brown, N, Marsden, B, von Delft, F, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-08-05 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | A fast, parallel method for efficiently exploring crystallization behaviour of large numbers of protein variants
To Be Published
|
|
9IYE
 
 | | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.39 A resolution. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, PHOSPHONOACETIC ACID, ... | | Authors: | Ahmad, N, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2024-07-30 | | Release date: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structure of Phosphopantetheine adenylyltransferase (PPAT) from Enterobacter spp. with the expression tag bound in the substrate binding site of a neighbouring molecule at 2.39 A resolution.
To Be Published
|
|
6UMM
 
 | | A complete structure of the ESX-3 translocon complex | | Descriptor: | ESX-3 secretion system ATPase EccB3, ESX-3 secretion system protein EccC3, ESX-3 secretion system protein EccD3, ... | | Authors: | Poweleit, N, Rosenberg, O.S. | | Deposit date: | 2019-10-09 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | A large inner membrane pore defines the ESX translocon
To Be Published
|
|
