7C75
 
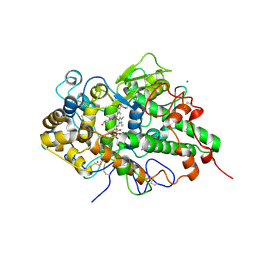 | | Crystal structure of yak lactoperoxidase with partially coordinated Na ion in the distal heme cavity | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Lactoperoxidase, ... | | Authors: | Singh, P.K, Viswanathan, V, Rani, C, Ahmad, N, Sharma, P, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Potassium-induced partial inhibition of lactoperoxidase: structure of the complex of lactoperoxidase with potassium ion at 2.20 angstrom resolution.
J.Biol.Inorg.Chem., 26, 2021
|
|
7CKR
 
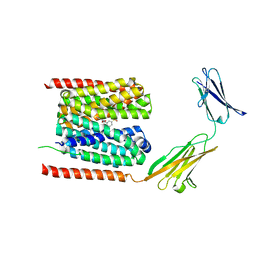 | | Cryo-EM structure of the human MCT1/Basigin-2 complex in the presence of anti-cancer drug candidate BAY-8002 in the outward-open conformation. | | Descriptor: | 2-[[2-chloranyl-5-(phenylsulfonyl)phenyl]carbonylamino]benzoic acid, Basigin, Monocarboxylate transporter 1 | | Authors: | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | Deposit date: | 2020-07-18 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
3L07
 
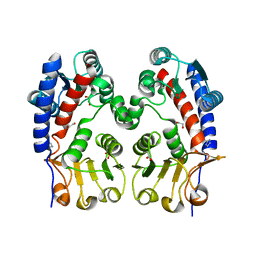 | | Methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase, putative bifunctional protein folD from Francisella tularensis. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Bifunctional protein folD, ... | | Authors: | Osipiuk, J, Maltseva, N, Mulligan, R, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-09 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | X-ray crystal structure of methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase, putative bifunctional protein folD from Francisella tularensis.
To be Published
|
|
7C74
 
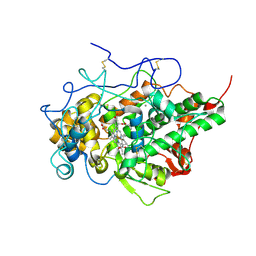 | | Crystal structure of yak lactoperoxidase using data obtained from crystals soaked in CaCl2 at 2.73 A resolution | | Descriptor: | CALCIUM ION, CHLORIDE ION, Lactoperoxidase, ... | | Authors: | Singh, P.K, Viswanathan, V, Pandey, S.N, Ahmad, N, Rani, C, Sharma, P, Sharma, P, Singh, T.P. | | Deposit date: | 2020-05-22 | | Release date: | 2020-06-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Potassium-induced partial inhibition of lactoperoxidase: structure of the complex of lactoperoxidase with potassium ion at 2.20 angstrom resolution.
J.Biol.Inorg.Chem., 26, 2021
|
|
7CD7
 
 | | GFP-40/GFPuv complex, Form I | | Descriptor: | GFP-40, Green fluorescent protein | | Authors: | Yasui, N, Yamashita, A. | | Deposit date: | 2020-06-18 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | A sweet protein monellin as a non-antibody scaffold for synthetic binding proteins.
J.Biochem., 169, 2021
|
|
3KQM
 
 | | Crystal Structure of hPNMT in Complex AdoHcy and 4-Bromo-1H-imidazole | | Descriptor: | 4-bromo-1H-imidazole, Phenylethanolamine N-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Drinkwater, N, Martin, J.L. | | Deposit date: | 2009-11-17 | | Release date: | 2010-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fragment-based screening by X-ray crystallography, MS and isothermal titration calorimetry to identify PNMT (phenylethanolamine N-methyltransferase) inhibitors.
Biochem.J., 431, 2010
|
|
7CN0
 
 | | Cryo-EM structure of K+-bound hERG channel | | Descriptor: | POTASSIUM ION, potassium channel 1 | | Authors: | Asai, T, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Senda, T, Murata, T. | | Deposit date: | 2020-07-29 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM Structure of K + -Bound hERG Channel Complexed with the Blocker Astemizole.
Structure, 29, 2021
|
|
7CKJ
 
 | | Crystal structure of CMP kinase in complex with CMP from Thermus thermophilus HB8 | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Cytidylate kinase | | Authors: | Mega, R, Nakagawa, N, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2020-07-17 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structures of Thermus thermophilus CMP kinase complexed with a phosphoryl group acceptor and donor.
PLoS ONE, 15, 2020
|
|
7CP0
 
 | | Crystal Structure of double mutant Y115E Y117E human Secretory Glutaminyl Cyclase | | Descriptor: | 1,2-ETHANEDIOL, Glutaminyl-peptide cyclotransferase, SULFATE ION, ... | | Authors: | Dileep, K.V, Ihara, K, Sakai, N, Shirozu, M, Zhang, K.Y.J. | | Deposit date: | 2020-08-05 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Piperidine-4-carboxamide as a new scaffold for designing secretory glutaminyl cyclase inhibitors.
Int.J.Biol.Macromol., 170, 2020
|
|
7CHK
 
 | | Cryo-EM Structure of Apple Latent Spherical Virus (ALSV) | | Descriptor: | VP20 protein, VP24 protein, VP25 protein | | Authors: | Naitow, H, Hamaguchi, T, Maki-Yonekura, S, Isogai, M, Yoshikawa, N, Yonekura, K. | | Deposit date: | 2020-07-06 | | Release date: | 2020-11-04 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Apple latent spherical virus structure with stable capsid frame supports quasi-stable protrusions expediting genome release.
Commun Biol, 3, 2020
|
|
3KT8
 
 | | Crystal structure of S. cerevisiae tryptophanyl-tRNA synthetase in complex with L-tryptophanamide | | Descriptor: | L-TRYPTOPHANAMIDE, SULFATE ION, Tryptophanyl-tRNA synthetase, ... | | Authors: | Zhou, M, Dong, X, Zhong, C, Shen, N, Ding, J. | | Deposit date: | 2009-11-24 | | Release date: | 2010-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structures of Saccharomyces cerevisiae tryptophanyl-tRNA synthetase: new insights into the mechanism of tryptophan activation and implications for anti-fungal drug design
Nucleic Acids Res., 38, 2010
|
|
7CKO
 
 | | Cryo-EM structure of the human MCT1/Basigin-2 complex in the presence of anti-cancer drug candidate 7ACC2 in the inward-open conformation | | Descriptor: | 7-[methyl-(phenylmethyl)amino]-2-oxidanylidene-chromene-3-carboxylic acid, Basigin, Monocarboxylate transporter 1 | | Authors: | Wang, N, Jiang, X, Zhang, S, Zhu, A, Yuan, Y, Lei, J, Yan, C. | | Deposit date: | 2020-07-18 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates.
Cell, 184, 2021
|
|
3L45
 
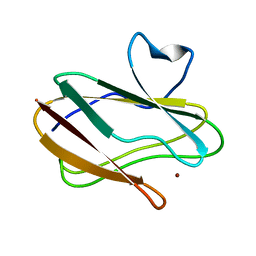 | | A Joint Neutron and X-ray structure of Oxidized Amicyanin | | Descriptor: | Amicyanin, COPPER (II) ION | | Authors: | Sukumar, N, Mathews, F.S, Langan, P, Davidson, V.L. | | Deposit date: | 2009-12-18 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-13 | | Method: | NEUTRON DIFFRACTION (1.8 Å), X-RAY DIFFRACTION | | Cite: | A joint x-ray and neutron study on amicyanin reveals the role of protein dynamics in electron transfer.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7C7D
 
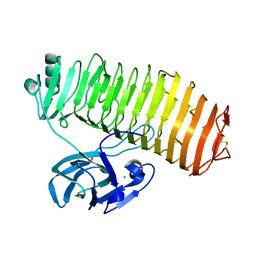 | | Crystal structure of the catalytic unit of thermostable GH87 alpha-1,3-glucanase from Streptomyces thermodiastaticus strain HF3-3 | | Descriptor: | CALCIUM ION, PENTAETHYLENE GLYCOL, alpha-1,3-glucanase | | Authors: | Itoh, T, Panti, N, Toyotake, Y, Hayashi, J, Suyotha, W, Yano, S, Wakayama, M, Hibi, T. | | Deposit date: | 2020-05-25 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Crystal structure of the catalytic unit of thermostable GH87 alpha-1,3-glucanase from Streptomyces thermodiastaticus strain HF3-3.
Biochem.Biophys.Res.Commun., 533, 2020
|
|
7CD8
 
 | | GFP-40/GFPuv complex, Form II | | Descriptor: | GFP-40, Green fluorescent protein | | Authors: | Yasui, N, Yamashita, A. | | Deposit date: | 2020-06-18 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A sweet protein monellin as a non-antibody scaffold for synthetic binding proteins.
J.Biochem., 169, 2021
|
|
7CJU
 
 | |
7CPO
 
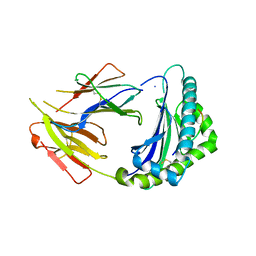 | | Crystal Structure of Anolis carolinensis MHC I complex | | Descriptor: | HIS-VAL-TYR-GLY-PRO-LEU-LYS-PRO-ILE, MHC CLASS I ANTIGEN, beta2-microglobulin | | Authors: | Wang, Y, Qu, Z, Ma, L, Wei, X, Zhang, N, Xia, C. | | Deposit date: | 2020-08-07 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Crystal Structure of the MHC Class I (MHC-I) Molecule in the Green Anole Lizard Demonstrates the Unique MHC-I System in Reptiles.
J Immunol., 206, 2021
|
|
7CGV
 
 | | Full consensus L-threonine 3-dehydrogenase, FcTDH-IIYM (NAD+ bound form) | | Descriptor: | Artificial L-threonine 3-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Motoyama, T, Hiramatsu, N, Asano, Y, Nakano, S, Ito, S. | | Deposit date: | 2020-07-02 | | Release date: | 2020-10-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Protein Sequence Selection Method That Enables Full Consensus Design of Artificial l-Threonine 3-Dehydrogenases with Unique Enzymatic Properties.
Biochemistry, 59, 2020
|
|
7CN1
 
 | | Cryo-EM structure of K+-bound hERG channel in the presence of astemizole | | Descriptor: | POTASSIUM ION, potassium channel | | Authors: | Asai, T, Adachi, N, Moriya, T, Kawasaki, M, Suzuki, K, Senda, T, Murata, T. | | Deposit date: | 2020-07-29 | | Release date: | 2021-01-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM Structure of K + -Bound hERG Channel Complexed with the Blocker Astemizole.
Structure, 29, 2021
|
|
7CWU
 
 | | SARS-CoV-2 spike proteins trimer in complex with P17 and FC05 Fabs cocktail | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Wang, N. | | Deposit date: | 2020-08-31 | | Release date: | 2020-12-16 | | Last modified: | 2021-01-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure-based development of human antibody cocktails against SARS-CoV-2.
Cell Res., 31, 2021
|
|
3L93
 
 | | Phosphopantetheine adenylyltransferase from Yersinia pestis. | | Descriptor: | FORMIC ACID, Phosphopantetheine adenylyltransferase | | Authors: | Osipiuk, J, Maltseva, N, Makowska-grzyska, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-04 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | X-ray crystal structure of phosphopantetheine adenylyltransferase from Yersinia pestis.
To be Published
|
|
5HO5
 
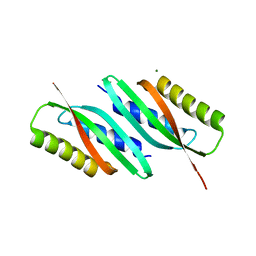 | | MamB | | Descriptor: | MAGNESIUM ION, Magnetosome protein MamB | | Authors: | Zeytuni, N, Keren, N, Davidov, G, Zarivach, R. | | Deposit date: | 2016-01-19 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The dual role of MamB in magnetosome membrane assembly and magnetite biomineralization.
Mol. Microbiol., 107, 2018
|
|
7C3G
 
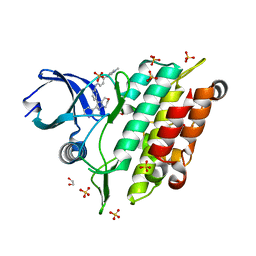 | | Crystal structure of human ALK2 kinase domain with R206H mutation in complex with a bicyclic pyrazole inhibitor RK-73134 | | Descriptor: | 1,2-ETHANEDIOL, Activin receptor type-1, SULFATE ION, ... | | Authors: | Sakai, N, Mishima-Tsumagari, C, Matsumoto, T, Shirouzu, M. | | Deposit date: | 2020-05-12 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Novel bicyclic pyrazoles as potent ALK2 (R206H) inhibitors for the treatment of fibrodysplasia ossificans progressiva.
Bioorg.Med.Chem.Lett., 38, 2021
|
|
3L6Y
 
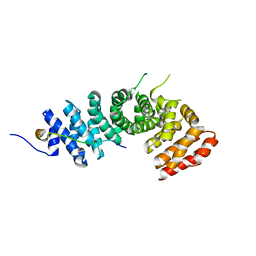 | | Crystal structure of p120 catenin in complex with E-cadherin | | Descriptor: | Catenin delta-1, E-cadherin | | Authors: | Ishiyama, N, Lee, S.-H, Liu, S, Li, G.-Y, Smith, M.J, Reichardt, L.F, Ikura, M. | | Deposit date: | 2009-12-27 | | Release date: | 2010-04-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion.
Cell(Cambridge,Mass.), 141, 2010
|
|
7COY
 
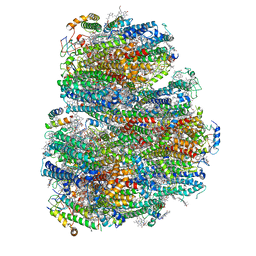 | | Structure of the far-red light utilizing photosystem I of Acaryochloris marina | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, CHLOROPHYLL D, ... | | Authors: | Kawakami, K, Yonekura, K, Hamaguchi, T, Kashino, Y, Shinzawa-Itoh, K, Inoue-Kashino, N, Itoh, S, Ifuku, K, Yamashita, E. | | Deposit date: | 2020-08-05 | | Release date: | 2021-03-31 | | Last modified: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of the far-red light utilizing photosystem I of Acaryochloris marina.
Nat Commun, 12, 2021
|
|
