8E0E
 
 | | nbF3:CaV beta subunit 2a complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Nirwan, N, Minor, D.L. | | Deposit date: | 2022-08-09 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selective posttranslational inhibition of Ca V beta 1 -associated voltage-dependent calcium channels with a functionalized nanobody.
Nat Commun, 13, 2022
|
|
7P9T
 
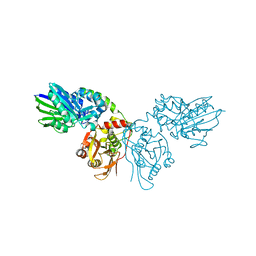 | | Crystal structure of CD73 in complex with dCMP in the open form | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 5'-nucleotidase, CALCIUM ION, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-07-27 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
6UAH
 
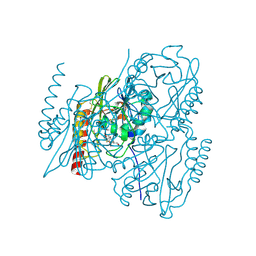 | | Crystal Structure of the Metallo-beta-Lactamase L1 from Stenotrophomonas maltophilia in the Complex with Hydrolyzed Meropenem | | Descriptor: | (2~{S},3~{R},4~{S})-2-[(2~{S},3~{R})-1,3-bis(oxidanyl)-1-oxidanylidene-butan-2-yl]-4-[(3~{S},5~{S})-5-(dimethylcarbamoy l)pyrrolidin-3-yl]sulfanyl-3-methyl-3,4-dihydro-2~{H}-pyrrole-5-carboxylic acid, 1,2-ETHANEDIOL, Putative metallo-beta-lactamase l1 (Beta-lactamase type ii) (Ec 3.5.2.6) (Penicillinase), ... | | Authors: | Kim, Y, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-09-10 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of the Metallo-beta-Lactamase L1 from Stenotrophomonas maltophilia in the Complex with Hydrolyzed Meropenem
To Be Published
|
|
8E34
 
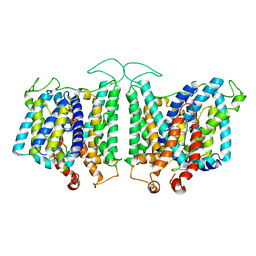 | | CryoEM structures of bAE1 captured in multiple states | | Descriptor: | Anion exchange protein | | Authors: | Zhekova, H.R, Wang, W.G, Jiang, J.S, Tsirulnikov, K, Muhammad-Khan, G.H, Azimov, R, Abuladze, N, Kao, L, Newman, D, Noskov, S.Y, Tieleman, P, Zhou, Z.H, Pushkin, A, Kurtz, I. | | Deposit date: | 2022-08-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | CryoEM structures of anion exchanger 1 capture multiple states of inward- and outward-facing conformations.
Commun Biol, 5, 2022
|
|
7PA4
 
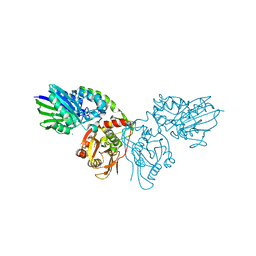 | | Crystal structure of CD73 in complex with CMP in the open form | | Descriptor: | 5'-nucleotidase, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-07-28 | | Release date: | 2021-10-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
6UBM
 
 | | AAV8 Baculovirus-Sf9 produced, empty capsid | | Descriptor: | Capsid protein | | Authors: | Paulk, N.K, Poweleit, N. | | Deposit date: | 2019-09-12 | | Release date: | 2020-06-03 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Methods Matter: Standard Production Platforms for Recombinant AAV Produce Chemically and Functionally Distinct Vectors.
To Be Published, 18, 2020
|
|
7P9N
 
 | | Crystal structure of CD73 in complex with AMP in the open form | | Descriptor: | 5'-nucleotidase, ADENOSINE MONOPHOSPHATE, CALCIUM ION, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-07-27 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
6EP3
 
 | | Lar controls the expression of the Listeria monocytogenes agr system and mediates virulence. | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Lmo0651 protein, ... | | Authors: | Pinheiro, J, Lisboa, J, Carvalho, F, Carreaux, A, Santos, N, Cabral, J, Sousa, S, Cabanes, D. | | Deposit date: | 2017-10-10 | | Release date: | 2018-07-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | MouR controls the expression of the Listeria monocytogenes Agr system and mediates virulence.
Nucleic Acids Res., 46, 2018
|
|
8E10
 
 | | Structure of mouse polymerase beta | | Descriptor: | DNA polymerase beta, MALONIC ACID | | Authors: | Thompson, M.K, Sharma, N, Prakash, A. | | Deposit date: | 2022-08-09 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Pol beta /XRCC1 heterodimerization dictates DNA damage recognition and basal Pol beta protein levels without interfering with mouse viability or fertility.
DNA Repair (Amst), 123, 2023
|
|
8E11
 
 | | Structure of mouse DNA polymerase Beta (PolB) mutant | | Descriptor: | ACETATE ION, DNA polymerase beta, MALONIC ACID | | Authors: | Sharma, N, Thompson, M.K, Prakash, A. | | Deposit date: | 2022-08-09 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Pol beta /XRCC1 heterodimerization dictates DNA damage recognition and basal Pol beta protein levels without interfering with mouse viability or fertility.
DNA Repair (Amst), 123, 2023
|
|
6UI6
 
 | | HBV T=3 149C3A | | Descriptor: | Core protein | | Authors: | Wu, W, Watts, N.R, Cheng, N, Huang, R, Steven, A, Wingfield, P.T. | | Deposit date: | 2019-09-30 | | Release date: | 2019-11-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Expression of quasi-equivalence and capsid dimorphism in the Hepadnaviridae.
Plos Comput.Biol., 16, 2020
|
|
7P9R
 
 | | Crystal structure of CD73 in complex with GMP in the open form | | Descriptor: | 5'-nucleotidase, CALCIUM ION, GLYCEROL, ... | | Authors: | Scaletti, E.R, Strater, N. | | Deposit date: | 2021-07-27 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Substrate binding modes of purine and pyrimidine nucleotides to human ecto-5'-nucleotidase (CD73) and inhibition by their bisphosphonic acid derivatives.
Purinergic Signal, 17, 2021
|
|
6E5C
 
 | | Solution NMR structure of a de novo designed double-stranded beta-helix | | Descriptor: | De novo beta protein | | Authors: | Marcos, E, Chidyausiku, T.M, McShan, A, Evangelidis, T, Nerli, S, Sgourakis, N, Tripsianes, K, Baker, D. | | Deposit date: | 2018-07-19 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | De novo design of a non-local beta-sheet protein with high stability and accuracy.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6UMQ
 
 | | Structure of DUF89 | | Descriptor: | Damage-control phosphatase DUF89, MAGNESIUM ION | | Authors: | Perry, J.J, Kenjic, N, Dennis, T.N. | | Deposit date: | 2019-10-10 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Human ARMT1 structure and substrate specificity indicates that it is a DUF89 family damage-control phosphatase.
J.Struct.Biol., 212, 2020
|
|
6UE9
 
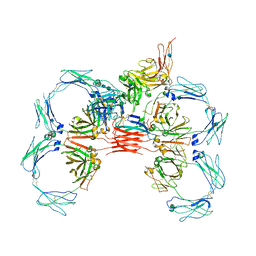 | | Structure of tetrameric sIgA complex (Class 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, ... | | Authors: | Kumar, N, Arthur, C.P, Ciferri, C, Matsumoto, M.L. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the secretory immunoglobulin A core.
Science, 367, 2020
|
|
6UE8
 
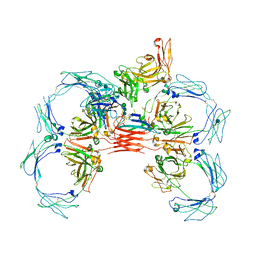 | | Structure of tetrameric sIgA complex (Class 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, ... | | Authors: | Kumar, N, Arthur, C.P, Ciferri, C, Matsumoto, M.L. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the secretory immunoglobulin A core.
Science, 367, 2020
|
|
1SQJ
 
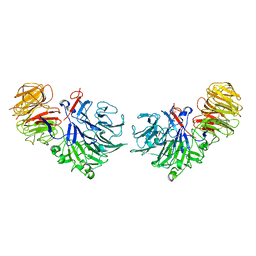 | | Crystal Structure Analysis of Oligoxyloglucan reducing-end-specific cellobiohydrolase (OXG-RCBH) | | Descriptor: | oligoxyloglucan reducing-end-specific cellobiohydrolase | | Authors: | Yaoi, K, Kondo, H, Noro, N, Suzuki, M, Tsuda, S, Mitsuishi, Y. | | Deposit date: | 2004-03-19 | | Release date: | 2004-07-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tandem Repeat of a Seven-Bladed beta-Propeller Domain in Oligoxyloglucan Reducing-End-Specific Cellobiohydrolase
Structure, 12, 2004
|
|
7TWU
 
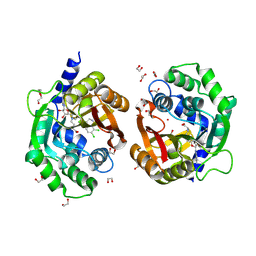 | | Crystal structure of human phenylethanolamine N-methyltransferase (PNMT) in complex with (2S)-2-amino-4-(((5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)(4-(7,8-dichloro-1,2,3,4-tetrahydroisoquinolin-4-yl)butyl)amino)butanoic acid and AdoHcy (SAH) | | Descriptor: | 1,2-ETHANEDIOL, 5'-([(3S)-3-amino-3-carboxypropyl]{4-[(4R)-7,8-dichloro-1,2,3,4-tetrahydroisoquinolin-4-yl]butyl}amino)-5'-deoxyadenosine, CADMIUM ION, ... | | Authors: | Harijan, R.K, Mahmoodi, N, Minnow, Y.V.T, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2022-02-07 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cell-Effective Transition-State Analogue of Phenylethanolamine N -Methyltransferase.
Biochemistry, 62, 2023
|
|
7TX2
 
 | | Crystal structure of human phenylethanolamine N-methyltransferase (PNMT) in complex with (2S)-2-amino-4-(((5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)(4-(7,8-dichloro-1,2,3,4-tetrahydroisoquinolin-4-yl)butyl)amino)butanoic acid | | Descriptor: | 1,2-ETHANEDIOL, 5'-([(3S)-3-amino-3-carboxypropyl]{4-[(4R)-7,8-dichloro-1,2,3,4-tetrahydroisoquinolin-4-yl]butyl}amino)-5'-deoxyadenosine, Phenylethanolamine N-methyltransferase | | Authors: | Harijan, R.K, Mahmoodi, N, Minnow, Y.V.T, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2022-02-07 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Cell-Effective Transition-State Analogue of Phenylethanolamine N -Methyltransferase.
Biochemistry, 62, 2023
|
|
6UJI
 
 | |
7P47
 
 | | Structure of the E3 ligase Smc5/Nse2 in complex with Ubc9-SUMO thioester mimetic | | Descriptor: | E3 SUMO-protein ligase MMS21, SUMO-conjugating enzyme UBC9, Structural maintenance of chromosomes protein 5, ... | | Authors: | Lascorz, J, Varejao, N, Reverter, D. | | Deposit date: | 2021-07-09 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.314 Å) | | Cite: | Structural basis for the E3 ligase activity enhancement of yeast Nse2 by SUMO-interacting motifs.
Nat Commun, 12, 2021
|
|
6UGM
 
 | | Structural basis of COMPASS eCM recognition of an unmodified nucleosome | | Descriptor: | Bre2, DNA (146-MER), H3 N-terminus, ... | | Authors: | Hsu, P.L, Shi, H, Zheng, N. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Basis of H2B Ubiquitination-Dependent H3K4 Methylation by COMPASS.
Mol.Cell, 76, 2019
|
|
7OVB
 
 | | L. pneumophila Type IV Coupling Complex (T4CC) with density for DotY N-terminal and middle domains | | Descriptor: | DotY, DotZ, IcmJ (DotN), ... | | Authors: | Mace, K, Meir, A, Lukoyanova, N, Waksman, G. | | Deposit date: | 2021-06-14 | | Release date: | 2021-12-01 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Proteins DotY and DotZ modulate the dynamics and localization of the type IVB coupling complex of Legionella pneumophila.
Mol.Microbiol., 117, 2022
|
|
6UMR
 
 | | Structure of DUF89 - D291A mutant | | Descriptor: | Damage-control phosphatase DUF89, MAGNESIUM ION | | Authors: | Perry, J.J, Kenjic, N, Dennis, T.N. | | Deposit date: | 2019-10-10 | | Release date: | 2020-07-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Human ARMT1 structure and substrate specificity indicates that it is a DUF89 family damage-control phosphatase.
J.Struct.Biol., 212, 2020
|
|
1SN6
 
 | |
