5ZML
 
 | | Stapled-peptides tailored against initiation of translation | | 分子名称: | 1,2-ETHANEDIOL, ACE-LYS-LYS-ARG-TYR-SER-ARG-MK8-GLN-LEU-LEU-MK8-PHE-ARG-ARG, Eukaryotic translation initiation factor 4E, ... | | 著者 | Lama, D, Liberator, A, Frosi, Y, Nakhle, J, Tsomia, N, Bashir, T, Lane, D.P, Brown, C.J, Verma, C.S, Auvin, S, Yano, J. | | 登録日 | 2018-04-04 | | 公開日 | 2019-02-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural insights reveal a recognition feature for tailoring hydrocarbon stapled-peptides against the eukaryotic translation initiation factor 4E protein.
Chem Sci, 10, 2019
|
|
6DR2
 
 | | Ca2+-bound human type 3 1,4,5-inositol trisphosphate receptor | | 分子名称: | CALCIUM ION, Inositol 1,4,5-trisphosphate receptor type 3, ZINC ION | | 著者 | Hite, R.K, Paknejad, N. | | 登録日 | 2018-06-11 | | 公開日 | 2018-07-18 | | 最終更新日 | 2018-08-15 | | 実験手法 | ELECTRON MICROSCOPY (4.33 Å) | | 主引用文献 | Structural basis for the regulation of inositol trisphosphate receptors by Ca2+and IP3.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6D4I
 
 | | Crystal Structure of a Fc Fragment of Rhesus macaque (Macaca mulatta) IgG2 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Fc fragment of IgG2 | | 著者 | Gohain, N, Tolbert, W.D, Pazgier, M. | | 登録日 | 2018-04-18 | | 公開日 | 2019-05-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | From Rhesus macaque to human: structural evolutionary pathways for immunoglobulin G subclasses.
Mabs, 11, 2019
|
|
6SHB
 
 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 1, in the presence of ssDNA | | 分子名称: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-08-06 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (3.07 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
5ZJY
 
 | | Stapled-peptides tailored against initiation of translation | | 分子名称: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E, LYS-LYS-ARG-TYR-SER-ARG-2JN-GLN-LEU-LEU-2JN-PHE | | 著者 | Lama, D, Liberator, A, Frosi, Y, Nakhle, J, Tsomia, N, Bashir, T, Lane, D.P, Brown, C.J, Verma, C.S, Auvin, S, Ciesielski, B, Uhring, M. | | 登録日 | 2018-03-22 | | 公開日 | 2019-03-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Structural insights reveal a recognition feature for tailoring hydrocarbon stapled-peptides against the eukaryotic translation initiation factor 4E protein.
Chem Sci, 10, 2019
|
|
5ZK5
 
 | | Stapled-peptides tailored against initiation of translation | | 分子名称: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E, LYS-ARG-TYR-SER-ARG-GLU-GLN-LEU-LEU-MK8-PHE-GLN-ARG-MK8 | | 著者 | Lama, D, Liberator, A, Frosi, Y, Nakhle, J, Tsomia, N, Bashir, T, Lane, D.P, Brown, C.J, Verma, C.S, Auvin, S, Ciesielski, F, Uhring, M. | | 登録日 | 2018-03-23 | | 公開日 | 2019-02-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Structural insights reveal a recognition feature for tailoring hydrocarbon stapled-peptides against the eukaryotic translation initiation factor 4E protein.
Chem Sci, 10, 2019
|
|
6DUW
 
 | |
6DV1
 
 | |
6DWK
 
 | | SAMHD1 Bound to Fludarabine-TP in the Catalytic Pocket | | 分子名称: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-fluoro-9-{5-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}phosphoryl]-beta-D-arabinofuranosyl}-9H-purin-6-a mine, ... | | 著者 | Knecht, K.M, Buzovetsky, O, Schneider, C, Thomas, D, Srikanth, V, Kaderali, L, Tofoleanu, F, Reiss, K, Ferreiros, N, Geisslinger, G, Batista, V.S, Ji, X, Cinatl, J, Keppler, O.T, Xiong, Y. | | 登録日 | 2018-06-26 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The structural basis for cancer drug interactions with the catalytic and allosteric sites of SAMHD1.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5YWE
 
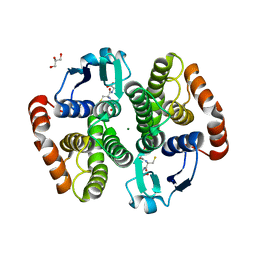 | | Crystal structure of hematopoietic prostaglandin D synthase apo form | | 分子名称: | GLUTATHIONE, GLYCEROL, Hematopoietic prostaglandin D synthase, ... | | 著者 | Kamo, M, Furubayashi, N, Inaka, K, Aritake, K, Urade, Y. | | 登録日 | 2017-11-29 | | 公開日 | 2018-12-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.68 Å) | | 主引用文献 | Crystal structure of hematopoietic prostaglandin D synthase apo form
To Be Published
|
|
6SHJ
 
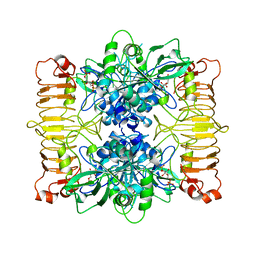 | | Escherichia coli AGPase in complex with FBP. Symmetry applied C2 | | 分子名称: | 1,6-di-O-phosphono-beta-D-fructofuranose, Glucose-1-phosphate adenylyltransferase | | 著者 | Cifuente, J.O, Comino, N, D'Angelo, C, Marina, A, Gil-Carton, D, Albesa-Jove, D, Guerin, M.E. | | 登録日 | 2019-08-07 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | The allosteric control mechanism of bacterial glycogen biosynthesis disclosed by cryoEM.
Curr Res Struct Biol, 2, 2020
|
|
6S8E
 
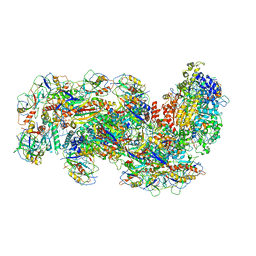 | | Cryo-EM structure of the type III-B Cmr-beta complex bound to non-cognate target RNA | | 分子名称: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-07-09 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
6BLK
 
 | |
6SH8
 
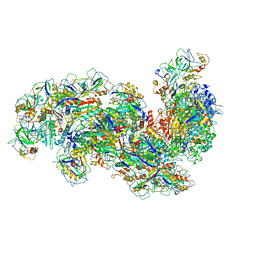 | | Cryo-EM structure of the Type III-B Cmr-beta bound to cognate target RNA and AMPPnP, state 2, in the presence of ssDNA | | 分子名称: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-08-06 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (3.14 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
7MCD
 
 | |
6BOZ
 
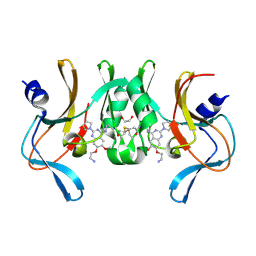 | | Structure of human SETD8 in complex with covalent inhibitor MS4138 | | 分子名称: | 1,2-ETHANEDIOL, N-(3-{[7-(2-aminoethoxy)-6-methoxy-2-(pyrrolidin-1-yl)quinazolin-4-yl]amino}propyl)prop-2-enamide, N-lysine methyltransferase KMT5A | | 著者 | Babault, N, Anqi, M, Jin, J. | | 登録日 | 2017-11-21 | | 公開日 | 2019-05-01 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | The dynamic conformational landscape of the protein methyltransferase SETD8.
Elife, 8, 2019
|
|
7MCC
 
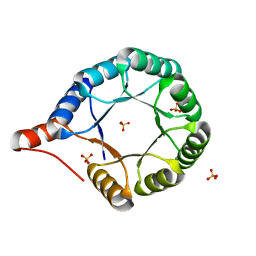 | | Crystal structure of an AI-designed TIM-barrel F2C | | 分子名称: | AI-designed TIM-barrel F2C, SULFATE ION | | 著者 | Mathews, I.I, Anand-Achim, N, Perez, C.P, Huang, P.S. | | 登録日 | 2021-04-02 | | 公開日 | 2022-01-19 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.46 Å) | | 主引用文献 | Protein sequence design with a learned potential.
Nat Commun, 13, 2022
|
|
6BMF
 
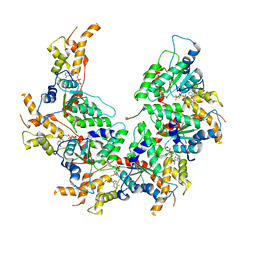 | | Vps4p-Vta1p complex with peptide binding to the central pore of Vps4p | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | 著者 | Han, H, Monroe, N, Shen, P, Sundquist, W.I, Hill, C.P. | | 登録日 | 2017-11-14 | | 公開日 | 2017-12-06 | | 実験手法 | ELECTRON MICROSCOPY (3.2 Å) | | 主引用文献 | The AAA ATPase Vps4 binds ESCRT-III substrates through a repeating array of dipeptide-binding pockets.
Elife, 6, 2017
|
|
6BUU
 
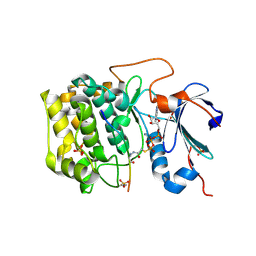 | | Crystal structure of AKT1 (aa 144-480) with a bisubstrate | | 分子名称: | GLY-ARG-PRO-ARG-THR-THR-ZXW-PHE-ALA-GLU, MANGANESE (II) ION, RAC-alpha serine/threonine-protein kinase, ... | | 著者 | Chu, N, Gabelli, S.B, Cole, P.A. | | 登録日 | 2017-12-11 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Akt Kinase Activation Mechanisms Revealed Using Protein Semisynthesis.
Cell, 174, 2018
|
|
3A9E
 
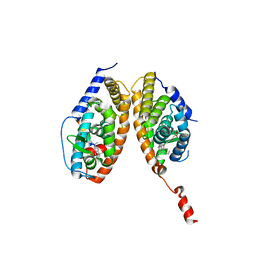 | | Crystal structure of a mixed agonist-bound RAR-alpha and antagonist-bound RXR-alpha heterodimer ligand binding domains | | 分子名称: | (2E,4E,6Z)-3-methyl-7-(5,5,8,8-tetramethyl-3-propoxy-5,6,7,8-tetrahydronaphthalen-2-yl)octa-2,4,6-trienoic acid, 13-mer (LXXLL motif) from Nuclear receptor coactivator 2, RETINOIC ACID, ... | | 著者 | Sato, Y, Duclaud, S, Peluso-Iltis, C, Poussin, P, Moras, D, Rochel, N, Structural Proteomics in Europe (SPINE) | | 登録日 | 2009-10-24 | | 公開日 | 2010-10-06 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | The Phantom Effect of the Rexinoid LG100754: structural and functional insights
Plos One, 5, 2010
|
|
1YHT
 
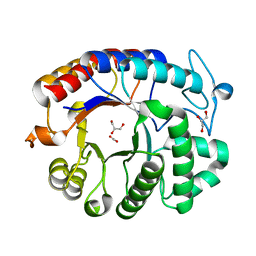 | | Crystal structure analysis of Dispersin B | | 分子名称: | ACETIC ACID, DspB, GLYCEROL | | 著者 | Ramasubbu, N, Thomas, L.M, Ragunath, C, Kaplan, J.B. | | 登録日 | 2005-01-10 | | 公開日 | 2006-01-10 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural Analysis of Dispersin B, a Biofilm-releasing Glycoside Hydrolase from the Periodontopathogen Actinobacillus actinomycetemcomitans.
J.Mol.Biol., 349, 2005
|
|
6CMC
 
 | |
6CRX
 
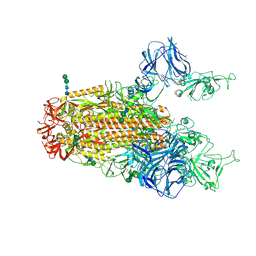 | | SARS Spike Glycoprotein, Stabilized variant, two S1 CTDs in the upwards conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | 著者 | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | 登録日 | 2018-03-19 | | 公開日 | 2018-04-11 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
6CUL
 
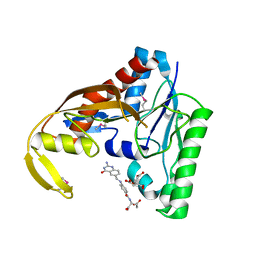 | | PvdF of pyoverdin biosynthesis is a structurally unique N10-formyltetrahydrofolate-dependent formyltransferase | | 分子名称: | CITRIC ACID, N-(4-{[(2-amino-4-oxo-1,4-dihydroquinazolin-6-yl)methyl]amino}benzene-1-carbonyl)-D-glutamic acid, Pyoverdine synthetase F | | 著者 | Kenjic, N, Hoag, M.R, Moraski, G.C, Caperelli, C.A, Moran, G.R, Lamb, A.L. | | 登録日 | 2018-03-26 | | 公開日 | 2019-02-06 | | 最終更新日 | 2019-11-27 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | PvdF of pyoverdin biosynthesis is a structurally unique N10-formyltetrahydrofolate-dependent formyltransferase.
Arch. Biochem. Biophys., 664, 2019
|
|
6CRW
 
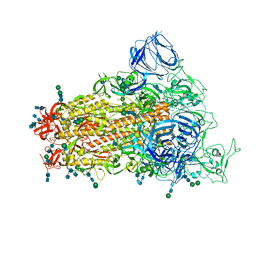 | | SARS Spike Glycoprotein, Stabilized variant, single upwards S1 CTD conformation | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin, ... | | 著者 | Kirchdoerfer, R.N, Wang, N, Pallesen, J, Turner, H.L, Cottrell, C.A, McLellan, J.S, Ward, A.B. | | 登録日 | 2018-03-19 | | 公開日 | 2018-04-11 | | 最終更新日 | 2020-07-29 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis.
Sci Rep, 8, 2018
|
|
