1D6P
 
 | | HUMAN LYSOZYME L63 MUTANT LABELLED WITH 2',3'-EPOXYPROPYL N,N'-DIACETYLCHITOBIOSE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, LYSOZYME | | Authors: | Muraki, M, Harata, K, Sugita, N, Sato, K. | | Deposit date: | 1999-10-15 | | Release date: | 2000-01-21 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Protein-carbohydrate interactions in human lysozyme probed by combining site-directed mutagenesis and affinity labeling.
Biochemistry, 39, 2000
|
|
7QYN
 
 | | Mus musculus acetylcholinesterase in complex with 2-((hydroxyimino)methyl)-1-(5-(4-methyl-3-nitrobenzamido)pentyl)pyridinium | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, 2-(2-METHOXYETHOXY)ETHANOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, ... | | Authors: | Forsgren, N, Lindgren, C, Edvinsson, L, Linusson, A, Ekstrom, F. | | Deposit date: | 2022-01-28 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Broad-Spectrum Antidote Discovery by Untangling the Reactivation Mechanism of Nerve-Agent-Inhibited Acetylcholinesterase.
Chemistry, 28, 2022
|
|
1OID
 
 | |
7R4E
 
 | | RVX-inhibited acetylcholinesterase in complex with 2-((hydroxyimino)methyl)-1-(5-(4-methyl-3-nitrobenzamido)pentyl)pyridinium | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, 2-(2-METHOXYETHOXY)ETHANOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, ... | | Authors: | Forsgren, N, Lindgren, C, Edvinsson, L, Linusson, A, Ekstrom, F. | | Deposit date: | 2022-02-08 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.00001168 Å) | | Cite: | Broad-Spectrum Antidote Discovery by Untangling the Reactivation Mechanism of Nerve-Agent-Inhibited Acetylcholinesterase.
Chemistry, 28, 2022
|
|
7R2F
 
 | | Structure of tabun inhibited acetylcholinesterase in complex with 2-((hydroxyimino)methyl)-1-(5-(4-methyl-3-nitrobenzamido)pentyl)pyridinium | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, 2-(2-METHOXYETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Forsgren, N, Lindgren, C, Edvinsson, L, Linusson, A, Ekstrom, F. | | Deposit date: | 2022-02-04 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broad-Spectrum Antidote Discovery by Untangling the Reactivation Mechanism of Nerve-Agent-Inhibited Acetylcholinesterase.
Chemistry, 28, 2022
|
|
7R0A
 
 | | Structure of sarin phosphonylated acetylcholinesterase in complex with 2-((hydroxyimino)methyl)-1-(5-(4-methyl-3-nitrobenzamido)pentyl)pyridinium | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, 2-(2-METHOXYETHOXY)ETHANOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, ... | | Authors: | Forsgren, N, Lindgren, C, Edvinsson, L, Linusson, A, Ekstrom, F. | | Deposit date: | 2022-02-01 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Broad-Spectrum Antidote Discovery by Untangling the Reactivation Mechanism of Nerve-Agent-Inhibited Acetylcholinesterase.
Chemistry, 28, 2022
|
|
7R02
 
 | | Mus musculus acetylcholinesterase in complex with N-(3-(diethylamino)propyl)-4-methyl-3-nitrobenzamide | | Descriptor: | 1,2-ETHANEDIOL, 2-(2-METHOXYETHOXY)ETHANOL, 2-[2-[2-[2-[2-[2-(2-methoxyethoxy)ethoxy]ethoxy]ethoxy]ethoxy]ethoxy]ethanol, ... | | Authors: | Forsgren, N, Lindgren, C, Edvinsson, L, Linusson, A, Ekstrom, F. | | Deposit date: | 2022-02-01 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Broad-Spectrum Antidote Discovery by Untangling the Reactivation Mechanism of Nerve-Agent-Inhibited Acetylcholinesterase.
Chemistry, 28, 2022
|
|
7R3C
 
 | | VX-inhibited acetylcholinesterase in complex with 2-((hydroxyimino)methyl)-1-(5-(4-methyl-3-nitrobenzamido)pentyl)pyridinium | | Descriptor: | 2,5,8,11,14,17-HEXAOXANONADECAN-19-OL, 2-(2-METHOXYETHOXY)ETHANOL, 2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXYL, ... | | Authors: | Forsgren, N, Lindgren, C, Edvinsson, L, Linusson, A, Ekstrom, F. | | Deposit date: | 2022-02-07 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.40000415 Å) | | Cite: | Broad-Spectrum Antidote Discovery by Untangling the Reactivation Mechanism of Nerve-Agent-Inhibited Acetylcholinesterase.
Chemistry, 28, 2022
|
|
5BYQ
 
 | | Semisynthetic [FeFe]-hydrogenase CpI with oxodithiolato-bridged [2Fe] cofactor | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Esselborn, J, Muraki, N, Engelbrecht, V, Hofmann, E, Kurisu, G, Happe, T. | | Deposit date: | 2015-06-10 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | A structural view of synthetic cofactor integration into [FeFe]-hydrogenases.
Chem Sci, 7, 2016
|
|
7MLF
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) Covalently Bound to Compound C7 | | Descriptor: | 3C-like proteinase, N-(4-tert-butylphenyl)-2-chloro-N-[(1R)-2-(cyclohexylamino)-2-oxo-1-(pyridin-3-yl)ethyl]acetamide | | Authors: | Sharon, I, Stille, J, Tjutrins, J, Wang, G, Venegas, F.A, Hennecker, C, Rueda, A.M, Miron, C.E, Pinus, S, Labarre, A, Patrascu, M.B, Vlaho, D, Huot, M, Mittermaier, A.K, Moitessier, N, Schmeing, T.M. | | Deposit date: | 2021-04-28 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design, synthesis and in vitro evaluation of novel SARS-CoV-2 3CL pro covalent inhibitors.
Eur.J.Med.Chem., 229, 2021
|
|
7MLG
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (3CLpro/Mpro) Covalently Bound to Compound C63 | | Descriptor: | (2R)-2-[(4-tert-butylphenyl)(ethanesulfonyl)amino]-N-cyclohexyl-2-(pyridin-3-yl)acetamide, 3C-like proteinase | | Authors: | Sharon, I, Stille, J, Tjutrins, J, Wang, G, Venegas, F.A, Hennecker, C, Rueda, A.M, Miron, C.E, Pinus, S, Labarre, A, Patrascu, M.B, Vlaho, D, Huot, M, Mittermaier, A.K, Moitessier, N, Schmeing, T.M. | | Deposit date: | 2021-04-28 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design, synthesis and in vitro evaluation of novel SARS-CoV-2 3CL pro covalent inhibitors.
Eur.J.Med.Chem., 229, 2021
|
|
1MB3
 
 | | CRYSTAL STRUCTURE OF THE RESPONSE REGULATOR DIVK AT PH 8.5 IN COMPLEX WITH MG2+ | | Descriptor: | MAGNESIUM ION, cell division response regulator DivK | | Authors: | Guillet, V, Ohta, N, Cabantous, S, Newton, A, Samama, J.-P, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2002-08-02 | | Release date: | 2002-12-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Crystallographic and Biochemical Studies of DivK Reveal Novel Features of
an Essential Response Regulator in Caulobacter crescentus.
J.Biol.Chem., 277, 2002
|
|
2QFC
 
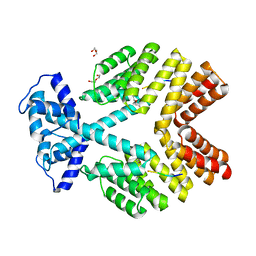 | | Crystal Structure of Bacillus thuringiensis PlcR complexed with PapR | | Descriptor: | C-terminus pentapeptide from PapR protein, DI(HYDROXYETHYL)ETHER, PlcR protein | | Authors: | Declerck, N, Chaix, D, Rugani, N, Hoh, F, Arold, S.T. | | Deposit date: | 2007-06-27 | | Release date: | 2007-11-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of PlcR: Insights into virulence regulation and evolution of quorum sensing in Gram-positive bacteria
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1J3B
 
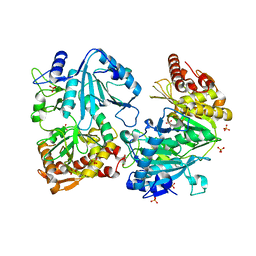 | | Crystal structure of ATP-dependent phosphoenolpyruvate carboxykinase from Thermus thermophilus HB8 | | Descriptor: | ATP-dependent phosphoenolpyruvate carboxykinase, CALCIUM ION, GLYCEROL, ... | | Authors: | Sugahara, M, Miyano, M, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-01-21 | | Release date: | 2003-02-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of ATP-dependent phosphoenolpyruvate carboxykinase from Thermus thermophilus HB8 showing the structural basis of induced fit and thermostability.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
7QYD
 
 | | mosquitocidal Cry11Ba determined at pH 6.5 from naturally-occurring nanocrystals by Serial femtosecond crystallography | | Descriptor: | Pesticidal crystal protein Cry11Ba | | Authors: | De Zitter, E, Tetreau, G, Andreeva, E.A, Coquelle, N, Colletier, J.-P, Sawaya, M.R, Schibrowsky, N.A, Cascio, D, Rodriguez, J.A. | | Deposit date: | 2022-01-28 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | De novo determination of mosquitocidal Cry11Aa and Cry11Ba structures from naturally-occurring nanocrystals.
Nat Commun, 13, 2022
|
|
1IPD
 
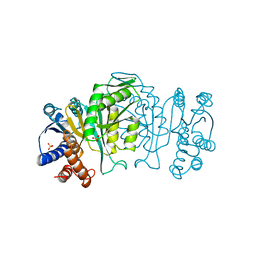 | | THREE-DIMENSIONAL STRUCTURE OF A HIGHLY THERMOSTABLE ENZYME, 3-ISOPROPYLMALATE DEHYDROGENASE OF THERMUS THERMOPHILUS AT 2.2 ANGSTROMS RESOLUTION | | Descriptor: | 3-ISOPROPYLMALATE DEHYDROGENASE, SULFATE ION | | Authors: | Imada, K, Sato, M, Tanaka, N, Katsube, Y, Matsuura, Y, Oshima, T. | | Deposit date: | 1992-01-29 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Three-dimensional structure of a highly thermostable enzyme, 3-isopropylmalate dehydrogenase of Thermus thermophilus at 2.2 A resolution.
J.Mol.Biol., 222, 1991
|
|
1IQW
 
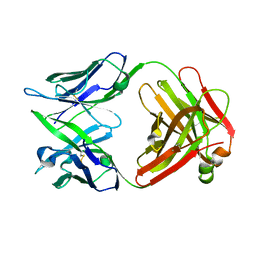 | | CRYSTAL STRUCTURE OF THE FAB FRAGMENT OF THE MOUSE ANTI-HUMAN FAS ANTIBODY HFE7A | | Descriptor: | ANTIBODY M-HFE7A, HEAVY CHAIN, LIGHT CHAIN | | Authors: | Ito, S, Takayama, T, Hanzawa, H, Ichikawa, K, Ohsumi, J, Serizawa, N, Hata, T, Haruyama, H. | | Deposit date: | 2001-08-10 | | Release date: | 2002-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the antigen-binding fragment of apoptosis-inducing mouse anti-human Fas monoclonal antibody HFE7A.
J.Biochem., 131, 2002
|
|
5BYS
 
 | | Semisynthetic [FeFe]-hydrogenase CpI with sulfur-dithiolato-bridged [2Fe] cofactor | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, IRON/SULFUR CLUSTER, Iron hydrogenase 1, ... | | Authors: | Esselborn, J, Muraki, N, Engelbrecht, V, Hofmann, E, Kurisu, G, Happe, T. | | Deposit date: | 2015-06-10 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A structural view of synthetic cofactor integration into [FeFe]-hydrogenases.
Chem Sci, 7, 2016
|
|
4YM5
 
 | | Crystal structure of the human nucleosome containing 6-4PP (inside) | | Descriptor: | 144 mer-DNA, 144-mer DNA, Histone H2A type 1-B/E, ... | | Authors: | Osakabe, A, Tachiwana, H, Kagawa, W, Horikoshi, N, Matsumoto, S, Hasegawa, M, Matsumoto, N, Toga, T, Yamamoto, J, Hanaoka, F, Thoma, N.H, Sugasawa, K, Iwai, S, Kurumizaka, H. | | Deposit date: | 2015-03-06 | | Release date: | 2015-12-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.005 Å) | | Cite: | Structural basis of pyrimidine-pyrimidone (6-4) photoproduct recognition by UV-DDB in the nucleosome
Sci Rep, 5, 2015
|
|
2QJT
 
 | | Crystal structure of a bifunctional NMN adenylyltransferase/ADP ribose pyrophosphatase complexed with AMP and MN ion from Francisella tularensis | | Descriptor: | ADENOSINE MONOPHOSPHATE, MANGANESE (II) ION, Nicotinamide-nucleotide adenylyltransferase | | Authors: | Huang, N, Sorci, L, Zhang, X, Brautigan, C, Raffaelli, N, Magni, G, Grishin, N.V, Osterman, A, Zhang, H. | | Deposit date: | 2007-07-09 | | Release date: | 2008-03-04 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bifunctional NMN Adenylyltransferase/ADP-Ribose Pyrophosphatase: Structure and Function in Bacterial NAD Metabolism.
Structure, 16, 2008
|
|
2H0Q
 
 | |
4X3B
 
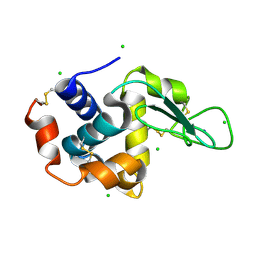 | | A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Roedig, P, Vartiainen, I, Duman, R, Panneerselvam, S, Stuebe, N, Lorbeer, O, Warmer, M, Sutton, G, Stuart, D.I, Weckert, E, David, C, Wagner, A, Meents, A. | | Deposit date: | 2014-11-28 | | Release date: | 2015-06-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A micro-patterned silicon chip as sample holder for macromolecular crystallography experiments with minimal background scattering.
Sci Rep, 5, 2015
|
|
4TNJ
 
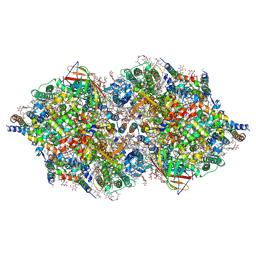 | | RT XFEL structure of Photosystem II 500 ms after the 2nd illumination (2F) at 4.5 A resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Kern, J, Tran, R, Alonso-Mori, R, Koroidov, S, Echols, N, Hattne, J, Ibrahim, M, Gul, S, Laksmono, H, Sierra, R.G, Gildea, R.J, Han, G, Hellmich, J, Lassalle-Kaiser, B, Chatterjee, R, Brewster, A, Stan, C.A, Gloeckner, C, Lampe, A, DiFiore, D, Milathianaki, D, Fry, A.R, Seibert, M.M, Koglin, J.E, Gallo, E, Uhlig, J, Sokaras, D, Weng, T.-C, Zwart, P.H, Skinner, D.E, Bogan, M.J, Messerschmidt, M, Glatzel, P, Williams, G.J, Boutet, S, Adams, P.D, Zouni, A, Messinger, J, Sauter, N.K, Bergmann, U, Yano, J, Yachandra, V.K. | | Deposit date: | 2014-06-04 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Taking snapshots of photosynthetic water oxidation using femtosecond X-ray diffraction and spectroscopy.
Nat Commun, 5, 2014
|
|
8BEL
 
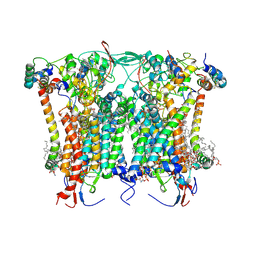 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CIII membrane domain) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-11 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.25 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
8BEH
 
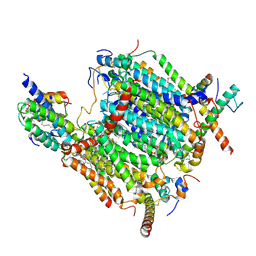 | | Cryo-EM structure of the Arabidopsis thaliana I+III2 supercomplex (CI membrane tip) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, ... | | Authors: | Klusch, N, Kuehlbrandt, W. | | Deposit date: | 2022-10-21 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Cryo-EM structure of the respiratory I + III 2 supercomplex from Arabidopsis thaliana at 2 angstrom resolution.
Nat.Plants, 9, 2023
|
|
