7UG8
 
 | | Crystal structure of a solute receptor from Synechococcus CC9311 in complex with alpha-ketovaleric and calcium | | 分子名称: | 1,2-ETHANEDIOL, 2-oxopentanoic acid, CALCIUM ION, ... | | 著者 | Shah, B.S, Mikolajek, H, Orr, C.M, Mykhaylyk, V, Owens, R.J, Paulsen, I.T. | | 登録日 | 2022-03-24 | | 公開日 | 2023-04-19 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.796 Å) | | 主引用文献 | Crystal structure of a solute receptor from Synechococcus CC9311 in complex with alpha-ketovaleric and calcium
To Be Published
|
|
5TCL
 
 | | Thaumatin from 4.96 A wavelength data collection | | 分子名称: | GLYCEROL, L(+)-TARTARIC ACID, Thaumatin-1 | | 著者 | Aurelius, O, Duman, R, El Omari, K, Mykhaylyk, V, Wagner, A. | | 登録日 | 2016-09-15 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (3.201 Å) | | 主引用文献 | Long-wavelength macromolecular crystallography - First successful native SAD experiment close to the sulfur edge.
Nucl Instrum Methods Phys Res B, 411, 2017
|
|
4ZG3
 
 | | In-vacuum long-wavelength crystallography | | 分子名称: | GLYCEROL, L(+)-TARTARIC ACID, SODIUM ION, ... | | 著者 | Wagner, A, Duman, R, Henderson, K, Mykhaylyk, V. | | 登録日 | 2015-04-22 | | 公開日 | 2016-03-09 | | 最終更新日 | 2016-03-23 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | In-vacuum long-wavelength macromolecular crystallography.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
8RF2
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 1E7 refined against the anomalous diffraction data | | 分子名称: | 1-benzothiophen-5-amine, Host translation inhibitor nsp1 | | 著者 | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | 登録日 | 2023-12-12 | | 公開日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RFC
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 7H2_AL1 refined against the anomalous diffraction data | | 分子名称: | (1~{R})-1-(4-bromophenyl)ethanamine, Host translation inhibitor nsp1 | | 著者 | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | 登録日 | 2023-12-12 | | 公開日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RFD
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 7H2_AL2 refined against the anomalous diffraction data | | 分子名称: | (1~{R})-1-(4-iodophenyl)ethanamine, Host translation inhibitor nsp1 | | 著者 | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | 登録日 | 2023-12-12 | | 公開日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.13 Å) | | 主引用文献 | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RF6
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 11A7_AL5 refined against the anomalous diffraction data | | 分子名称: | 6-iodanyl-2,3-dihydro-1,3-benzothiazol-2-amine, Host translation inhibitor nsp1 | | 著者 | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | 登録日 | 2023-12-12 | | 公開日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.08 Å) | | 主引用文献 | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RF5
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 11A7 refined against the anomalous diffraction data | | 分子名称: | 6-fluoro-1,3-benzothiazol-2-amine, Host translation inhibitor nsp1 | | 著者 | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | 登録日 | 2023-12-12 | | 公開日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RF3
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 7G3 refined against the anomalous diffraction data | | 分子名称: | 2-(1-benzothiophen-3-yl)ethanoic acid, Host translation inhibitor nsp1 | | 著者 | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | 登録日 | 2023-12-12 | | 公開日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.23 Å) | | 主引用文献 | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RFF
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 6A6 refined against the anomalous diffraction data | | 分子名称: | 1,3-benzothiazol-2-amine, Host translation inhibitor nsp1 | | 著者 | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | 登録日 | 2023-12-12 | | 公開日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.31 Å) | | 主引用文献 | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RF4
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 9D4 refined against the anomalous diffraction data | | 分子名称: | 4-chloranyl-1~{H}-indazol-3-amine, Host translation inhibitor nsp1 | | 著者 | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | 登録日 | 2023-12-12 | | 公開日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.11 Å) | | 主引用文献 | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RF8
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 11A7_AL6 refined against the anomalous diffraction data | | 分子名称: | 6-bromanyl-1,3-benzothiazol-2-amine, Host translation inhibitor nsp1 | | 著者 | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | 登録日 | 2023-12-12 | | 公開日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.12 Å) | | 主引用文献 | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
6YO1
 
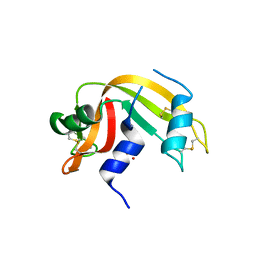 | | Crystal structure of ribonuclease A solved by vanadium SAD phasing | | 分子名称: | Ribonuclease pancreatic, URIDINE-2',3'-VANADATE | | 著者 | El Omari, K, Mohamad, N, Bountra, K, Duman, R, Romano, M, Schlegel, K, Kwong, H, Mykhaylyk, V, Olesen, C.E, Moller, J.V, Bublitz, M, Beis, K, Wagner, A. | | 登録日 | 2020-04-14 | | 公開日 | 2020-11-04 | | 最終更新日 | 2020-12-02 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Experimental phasing with vanadium and application to nucleotide-binding membrane proteins.
Iucrj, 7, 2020
|
|
6YSO
 
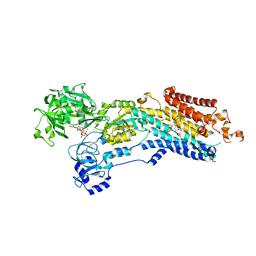 | | Crystal structure of the (SR) Ca2+-ATPase solved by vanadium SAD phasing | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, OCTANOIC ACID [3S-[3ALPHA, ... | | 著者 | El Omari, K, Mohamad, N, Bountra, K, Duman, R, Romano, M, Schlegel, K, Kwong, H, Mykhaylyk, V, Olesen, C.E, Moller, J.V, Bublitz, M, Beis, K, Wagner, A. | | 登録日 | 2020-04-22 | | 公開日 | 2020-11-04 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (3.13 Å) | | 主引用文献 | Experimental phasing with vanadium and application to nucleotide-binding membrane proteins.
Iucrj, 7, 2020
|
|
6OWY
 
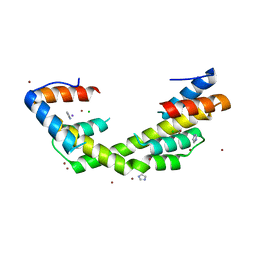 | | Spy H96L:Im7 K20pI-Phe complex; multiple anomalous datasets contained herein for element identification | | 分子名称: | CHLORIDE ION, IMIDAZOLE, IODIDE ION, ... | | 著者 | Rocchio, S, Duman, R, El Omari, K, Mykhaylyk, V, Yan, Z, Wagner, A, Bardwell, J.C.A, Horowitz, S. | | 登録日 | 2019-05-12 | | 公開日 | 2019-05-29 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Identifying dynamic, partially occupied residues using anomalous scattering.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6OWX
 
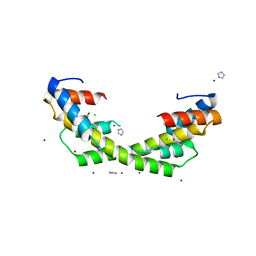 | | Spy H96L:Im7 L18pI-Phe complex; multiple anomalous datasets contained herein for element identification | | 分子名称: | CHLORIDE ION, IMIDAZOLE, IODIDE ION, ... | | 著者 | Rocchio, S, Duman, R, El Omari, K, Mykhaylyk, V, Yan, Z, Wagner, A, Bardwell, J.C.A, Horowitz, S. | | 登録日 | 2019-05-12 | | 公開日 | 2019-05-29 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.06 Å) | | 主引用文献 | Identifying dynamic, partially occupied residues using anomalous scattering.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6OWZ
 
 | | Spy H96L:Im7 L19pI-Phe complex; multiple anomalous datasets contained herein for element identification | | 分子名称: | CHLORIDE ION, IMIDAZOLE, IODIDE ION, ... | | 著者 | Rocchio, S, Duman, R, El Omari, K, Mykhaylyk, V, Yan, Z, Wagner, A, Bardwell, J.C.A, Horowitz, S. | | 登録日 | 2019-05-12 | | 公開日 | 2019-05-29 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Identifying dynamic, partially occupied residues using anomalous scattering.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
8CRF
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 5E11 refined against anomalous diffraction data | | 分子名称: | Host translation inhibitor nsp1, ~{N}-methyl-1-(4-thiophen-2-ylphenyl)methanamine | | 著者 | Ma, S, Mykhaylyk, V, Pinotsis, N, Bowler, M.W, Kozielski, F. | | 登録日 | 2023-03-08 | | 公開日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.15 Å) | | 主引用文献 | High-Confidence Placement of Fragments into Electron Density Using Anomalous Diffraction-A Case Study Using Hits Targeting SARS-CoV-2 Non-Structural Protein 1.
Int J Mol Sci, 24, 2023
|
|
7ZVX
 
 | | Crystal structure of human Annexin A2 in complex with full phosphorothioate 5-10 2'-methoxyethyl DNA gapmer antisense oligonucleotide solved at 2.4 A resolution | | 分子名称: | 1,2-ETHANEDIOL, 2'-methoxyethyl DNA gapmer antisense oligonucleotide, Annexin A2, ... | | 著者 | Hyjek-Skladanowska, M, Anderson, B, Mykhaylyk, V, Orr, C, Wagner, A, Skowronek, K, Seth, P, Nowotny, M. | | 登録日 | 2022-05-17 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Structures of annexin A2-PS DNA complexes show dominance of hydrophobic interactions in phosphorothioate binding.
Nucleic Acids Res., 51, 2023
|
|
7ZVN
 
 | | Crystal structure of human Annexin A2 in complex with full phosphorothioate 5-10 2'-methoxyethyl DNA gapmer antisense oligonucleotide solved at 1.87 A resolution | | 分子名称: | 2'-methoxyethyl DNA gapmer antisense oligonucleotide, Annexin A2, CALCIUM ION, ... | | 著者 | Hyjek-Skladanowska, M, Anderson, B, Mykhaylyk, V, Orr, C, Wagner, A, Skowronek, K, Seth, P, Nowotny, M. | | 登録日 | 2022-05-16 | | 公開日 | 2022-09-14 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | Structures of annexin A2-PS DNA complexes show dominance of hydrophobic interactions in phosphorothioate binding.
Nucleic Acids Res., 51, 2023
|
|
7S6E
 
 | | Crystal structure of UrtA from Synechococcus CC9311 in complex with urea and calcium | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | 著者 | Shah, B.S, Mikolajek, H, Mykhaylyk, V, Orr, C.M, Owens, R.J, Paulsen, I.T. | | 登録日 | 2021-09-14 | | 公開日 | 2021-10-13 | | 実験手法 | X-RAY DIFFRACTION (1.973 Å) | | 主引用文献 | Crystal structure of UrtA from Synechococcus CC9311 in complex with urea and calcium
To Be Published
|
|
7S6F
 
 | | Crystal structure of UrtA1 from Synechococcus WH8102 in complex with urea and calcium | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, Putative urea ABC transporter, ... | | 著者 | Shah, B.S, Mikolajek, H, Orr, C.M, Mykhaylyk, V, Owens, R.J, Paulsen, I.T. | | 登録日 | 2021-09-14 | | 公開日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of UrtA1 from Synechococcus WH8102 in complex with urea and calcium
To Be Published
|
|
7S6G
 
 | | Crystal structure of PhnD from Synechococcus MITS9220 in complex with phosphate | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, PHOSPHATE ION, ... | | 著者 | Shah, B.S, Mikolajek, H, Orr, C.M, Mykhaylyk, V, Owens, R.J, Paulsen, I.T. | | 登録日 | 2021-09-14 | | 公開日 | 2021-10-27 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Marine picocyanobacterial PhnD1 shows specificity for various phosphorus sources but likely represents a constitutive inorganic phosphate transporter.
Isme J, 17, 2023
|
|
8PXC
 
 | | Structure of Fap1, a domain of the accessory Sec-dependent serine-rich glycoprotein adhesin from Streptococcus oralis, solved at wavelength 3.06 A | | 分子名称: | 1,2-ETHANEDIOL, CALCIUM ION, Fap1 | | 著者 | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Owen, C.D, Walsh, M.A, Wagner, A. | | 登録日 | 2023-07-23 | | 公開日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.973 Å) | | 主引用文献 | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PX5
 
 | | Structure of the RNA recognition motif (RRM) of Seb1 from S. pombe., solved at wavelength 2.75 A | | 分子名称: | Rpb7-binding protein seb1 | | 著者 | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Wittmann, S, Renner, M, Grimes, J.M, Wagner, A. | | 登録日 | 2023-07-22 | | 公開日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.77 Å) | | 主引用文献 | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
