6CUL
 
 | | PvdF of pyoverdin biosynthesis is a structurally unique N10-formyltetrahydrofolate-dependent formyltransferase | | Descriptor: | CITRIC ACID, N-(4-{[(2-amino-4-oxo-1,4-dihydroquinazolin-6-yl)methyl]amino}benzene-1-carbonyl)-D-glutamic acid, Pyoverdine synthetase F | | Authors: | Kenjic, N, Hoag, M.R, Moraski, G.C, Caperelli, C.A, Moran, G.R, Lamb, A.L. | | Deposit date: | 2018-03-26 | | Release date: | 2019-02-06 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | PvdF of pyoverdin biosynthesis is a structurally unique N10-formyltetrahydrofolate-dependent formyltransferase.
Arch. Biochem. Biophys., 664, 2019
|
|
4IYF
 
 | |
4IYD
 
 | |
5KKC
 
 | | l-lactate dehydrogenase from rabbit muscle with the inhibitor 6DHNAD | | Descriptor: | L-lactate dehydrogenase A chain, SULFATE ION, [[(2~{R},3~{S},4~{R},5~{R})-5-(5-aminocarbonyl-2~{H}-pyridin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Meneely, K.M, Moran, G.R, Lamb, A.L. | | Deposit date: | 2016-06-21 | | Release date: | 2016-11-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Ligand binding phenomena that pertain to the metabolic function of renalase.
Arch.Biochem.Biophys., 612, 2016
|
|
5KRQ
 
 | | Renalase in complex with NADPH | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Renalase | | Authors: | Silvaggi, N.R, Moran, G.R, Roman, J.V. | | Deposit date: | 2016-07-07 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.086 Å) | | Cite: | Ligand binding phenomena that pertain to the metabolic function of renalase.
Arch.Biochem.Biophys., 612, 2016
|
|
4O53
 
 | | Crystal Structure of Trichomonas vaginalis Triosephosphate Isomerase Ile45-Leu mutant (Tvag_497370) | | Descriptor: | SODIUM ION, Triosephosphate isomerase | | Authors: | Lara-Gonzalez, S, Montero-Moran, G.M, Estrella-Hernandez, P, Benitez-Cardoza, C.G, Brieba, L.G. | | Deposit date: | 2013-12-19 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering mutants with altered dimer-monomer equilibrium reveal the existence of stable monomeric Triosephosphate isomerases
To be Published
|
|
4O52
 
 | |
4O57
 
 | | Crystal Structure of Trichomonas vaginalis Triosephosphate Isomerase Ile45-Tyr mutant (Tvag_497370) | | Descriptor: | SODIUM ION, Triosephosphate isomerase | | Authors: | Lara-Gonzalez, S, Estrella-Hernandez, P, Montero-Moran, G.M, Benitez-Cardoza, C.G, Brieba, L.G. | | Deposit date: | 2013-12-19 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.793 Å) | | Cite: | Engineering mutants with altered dimer-monomer equilibrium reveal the existence of stable monomeric Triosephosphate isomerases
To be Published
|
|
5KKA
 
 | | E. coli malate dehydrogenase with the inhibitor 6DHNAD | | Descriptor: | Malate dehydrogenase, [[(2~{R},3~{S},4~{R},5~{R})-5-(5-aminocarbonyl-2~{H}-pyridin-1-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Meneely, K.M, Lamb, A.L, Moran, G.R. | | Deposit date: | 2016-06-21 | | Release date: | 2016-11-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Ligand binding phenomena that pertain to the metabolic function of renalase.
Arch.Biochem.Biophys., 612, 2016
|
|
8F5W
 
 | | Dihydropyrimidine Dehydrogenase (DPD) C671S Mutant Soaked with Dihydrothymine and NADPH Quasi-Anaerobically | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, Dihydropyrimidine dehydrogenase [NADP(+)], FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kaley, N, Smith, M, Forouzesh, D, Liu, D, Moran, G. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mammalian dihydropyrimidine dehydrogenase: Added mechanistic details from transient-state analysis of charge transfer complexes.
Arch.Biochem.Biophys., 736, 2023
|
|
8F61
 
 | | Dihydropyrimidine Dehydrogenase (DPD) C671S Mutant Soaked with Dihydrothymine Quasi-Anaerobically | | Descriptor: | (5S)-5-methyl-1,3-diazinane-2,4-dione, 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, Dihydropyrimidine dehydrogenase [NADP(+)], ... | | Authors: | Kaley, N, Smith, M, Forouzesh, D, Liu, D, Moran, G. | | Deposit date: | 2022-11-15 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Mammalian dihydropyrimidine dehydrogenase: Added mechanistic details from transient-state analysis of charge transfer complexes.
Arch.Biochem.Biophys., 736, 2023
|
|
8F6N
 
 | | Dihydropyrimidine Dehydrogenase (DPD) C671S Mutant Soaked with Thymine Quasi-Anaerobically | | Descriptor: | Dihydropyrimidine dehydrogenase [NADP(+)], FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Kaley, N, Smith, M, Forouzesh, D, Liu, D, Moran, G. | | Deposit date: | 2022-11-16 | | Release date: | 2023-02-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Mammalian dihydropyrimidine dehydrogenase: Added mechanistic details from transient-state analysis of charge transfer complexes.
Arch.Biochem.Biophys., 736, 2023
|
|
6PAY
 
 | | Structure of HsICDH1:Mg(II):ICT:NADPH(50%) complex reveals structural basis for observation of half-sites reactivity | | Descriptor: | FORMIC ACID, ISOCITRIC ACID, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Silvaggi, N.R, Melkonian, T.R, Roman, J.V, Moran, G.R. | | Deposit date: | 2019-06-12 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Transient-State Analysis of Human Isocitrate Dehydrogenase I: Accounting for the Interconversion of Active and Non-Active Conformational States.
Biochemistry, 58, 2019
|
|
7JX9
 
 | | The crystal structure of human ornithine aminotransferase with an intermediate bound during inactivation by (1S,3S)-3-amino-4-(hexafluoropropan-2-ylidenyl)-cyclopentane-1-carboxylic acid. | | Descriptor: | (1S,3S,4S)-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-4-(1,1,3,3,3-pentafluoroprop-1-en-2-yl)cyclopentane-1-carboxylic acid, N-[1,3-dihydroxy-2-(hydroxymethyl)propan-2-yl]glycine, Ornithine aminotransferase, ... | | Authors: | Butrin, A, Beaupre, B, Shen, S, Silverman, R.B, Moran, G, Liu, D. | | Deposit date: | 2020-08-26 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and Kinetic Analyses Reveal the Dual Inhibition Modes of Ornithine Aminotransferase by (1 S ,3 S )-3-Amino-4-(hexafluoropropan-2-ylidenyl)-cyclopentane-1-carboxylic Acid (BCF 3 ).
Acs Chem.Biol., 16, 2021
|
|
2R5V
 
 | | Hydroxymandelate Synthase Crystal Structure | | Descriptor: | (2S)-hydroxy(4-hydroxyphenyl)ethanoic acid, COBALT (II) ION, PCZA361.1, ... | | Authors: | Brownlee, J.M, He, P, Moran, G.R, Harrison, D.H.T. | | Deposit date: | 2007-09-04 | | Release date: | 2008-03-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Two roads diverged: the structure of hydroxymandelate synthase from Amycolatopsis orientalis in complex with 4-hydroxymandelate.
Biochemistry, 47, 2008
|
|
4ZCD
 
 | | Renalase in complex with NAD+ | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Silvaggi, N.R, Moran, G.R, Roman, J.V. | | Deposit date: | 2015-04-15 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6605 Å) | | Cite: | Bacterial Renalase: Structure and Kinetics of an Enzyme with 2- and 6-Dihydro-beta-NAD(P) Oxidase Activity from Pseudomonas phaseolicola.
Biochemistry, 54, 2015
|
|
4ZCC
 
 | | Renalase in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | Authors: | Silvaggi, N.R, Moran, G.R, Roman, J.V. | | Deposit date: | 2015-04-15 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Bacterial Renalase: Structure and Kinetics of an Enzyme with 2- and 6-Dihydro-beta-NAD(P) Oxidase Activity from Pseudomonas phaseolicola.
Biochemistry, 54, 2015
|
|
7JH4
 
 | | Crystal structure of NAD(P)H-flavin oxidoreductase (NfoR) from S. aureus complexed with reduced FMN and NAD+ | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, NAD(P)H-dependent oxidoreductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Zheng, Y, O'Neill, A.G, Beaupre, B.A, Liu, D, Moran, G.R. | | Deposit date: | 2020-07-20 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | NfoR: Chromate Reductase or Flavin Mononucleotide Reductase?
Appl.Environ.Microbiol., 86, 2020
|
|
7LJT
 
 | | Porcine Dihydropyrimidine Dehydrogenase (DPD) soaked with 5-Ethynyluracil (5EU), NADPH - 20 minutes | | Descriptor: | 5-ethynylpyrimidine-2,4(1H,3H)-dione, Dihydropyrimidine dehydrogenase [NADP(+)], FLAVIN MONONUCLEOTIDE, ... | | Authors: | Butrin, A, Forouzesh, D, Beaupre, B, Wawrzak, Z, Liu, D, Moran, G. | | Deposit date: | 2021-01-30 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The Interaction of Porcine Dihydropyrimidine Dehydrogenase with the Chemotherapy Sensitizer: 5-Ethynyluracil.
Biochemistry, 60, 2021
|
|
7LJU
 
 | | Porcine Dihydropyrimidine Dehydrogenase (DPD) crosslinked with 5-Ethynyluracil (5EU) | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, 5-ethynylpyrimidine-2,4(1H,3H)-dione, Dihydropyrimidine dehydrogenase [NADP(+)], ... | | Authors: | Butrin, A, Forouzesh, D, Beaupre, B, Wawrzak, Z, Liu, D, Moran, G. | | Deposit date: | 2021-01-30 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The Interaction of Porcine Dihydropyrimidine Dehydrogenase with the Chemotherapy Sensitizer: 5-Ethynyluracil.
Biochemistry, 60, 2021
|
|
7M32
 
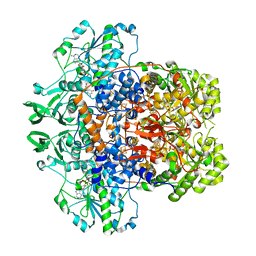 | | Dihydropyrimidine Dehydrogenase (DPD) C671A Mutant Soaked with Uracil and NADPH Anaerobically | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, ALANINE, Dihydropyrimidine dehydrogenase [NADP(+)], ... | | Authors: | Butrin, A, Beaupre, B, Forouzesh, D, Liu, D, Moran, G. | | Deposit date: | 2021-03-18 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Perturbing the Movement of Hydrogens to Delineate and Assign Events in the Reductive Activation and Turnover of Porcine Dihydropyrimidine Dehydrogenase.
Biochemistry, 60, 2021
|
|
7LJS
 
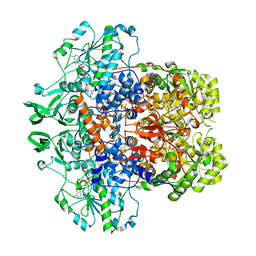 | | Porcine Dihydropyrimidine dehydrogenase (DPD) complexed with 5-Ethynyluracil (5EU) - Open Form | | Descriptor: | 5-ethynylpyrimidine-2,4(1H,3H)-dione, Dihydropyrimidine dehydrogenase [NADP(+)], FLAVIN MONONUCLEOTIDE, ... | | Authors: | Butrin, A, Forouzesh, D, Beaupre, B, Wawrzak, Z, Liu, D, Moran, G. | | Deposit date: | 2021-01-30 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Interaction of Porcine Dihydropyrimidine Dehydrogenase with the Chemotherapy Sensitizer: 5-Ethynyluracil.
Biochemistry, 60, 2021
|
|
7M31
 
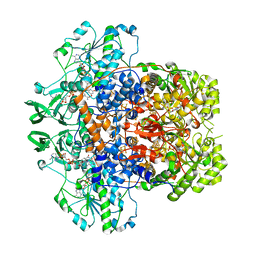 | | Dihydropyrimidine Dehydrogenase (DPD) C671S Mutant Soaked with Thymine and NADPH Anaerobically | | Descriptor: | Dihydropyrimidine dehydrogenase [NADP(+)], FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Butrin, A, Beaupre, B, Forouzesh, D, Liu, D, Moran, G. | | Deposit date: | 2021-03-18 | | Release date: | 2021-06-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Perturbing the Movement of Hydrogens to Delineate and Assign Events in the Reductive Activation and Turnover of Porcine Dihydropyrimidine Dehydrogenase.
Biochemistry, 60, 2021
|
|
1T47
 
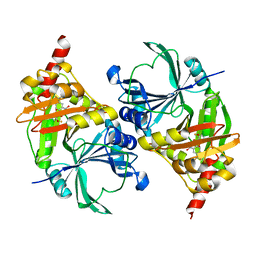 | | Structure of fe2-HPPD bound to NTBC | | Descriptor: | 2-{HYDROXY[2-NITRO-4-(TRIFLUOROMETHYL)PHENYL]METHYLENE}CYCLOHEXANE-1,3-DIONE, 4-hydroxyphenylpyruvate dioxygenase, FE (II) ION | | Authors: | Brownlee, J, Johnson-Winters, K, Harrison, D.H.T, Moran, G.R. | | Deposit date: | 2004-04-28 | | Release date: | 2004-06-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Ferrous Form of (4-Hydroxyphenyl)pyruvate Dioxygenase from Streptomyces avermitilis in Complex with the Therapeutic Herbicide, NTBC
Biochemistry, 43, 2004
|
|
