2PFV
 
 | |
2PFT
 
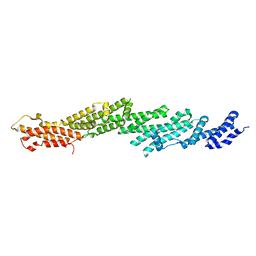 | |
2X43
 
 | | STRUCTURAL BASIS OF MOLECULAR RECOGNITION BY SHERP AT MEMBRANE SURFACES | | Descriptor: | SHERP | | Authors: | Moore, B, Miles, A.J, Guerra, C.G, Simpson, P, Iwata, M, Wallace, B.A, Matthews, S.J, Smith, D.F, Brown, K.A. | | Deposit date: | 2010-02-09 | | Release date: | 2010-11-24 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Moelcular Recognition by the Leishmania Small Hydrophilic Endoplasmic Reticulum-Associated Protein, Sherp, at Membrane Surfaces
J.Biol.Chem., 286, 2011
|
|
8FFT
 
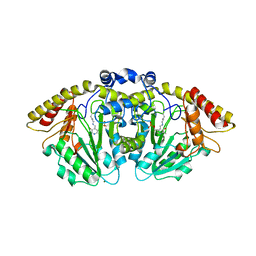 | | Structure of GntC, a PLP-dependent enzyme catalyzing L-enduracididine biosynthesis from (S)-4-hydroxy-L-arginine | | Descriptor: | Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, MAGNESIUM ION | | Authors: | Chen, P.Y.-T, Lima, S.T, Chekan, J.R, Moore, B.S. | | Deposit date: | 2022-12-10 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanistic and Structural Insights into a Divergent PLP-Dependent l-Enduracididine Cyclase from a Toxic Cyanobacterium.
Acs Catalysis, 13, 2023
|
|
8FFU
 
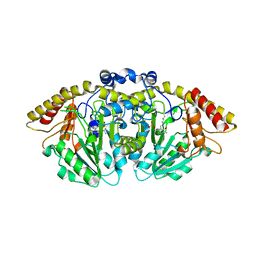 | | Structure of GntC, a PLP-dependent enzyme catalyzing L-enduracididine biosynthesis from (S)-4-hydroxy-L-arginine, with the substrate bound | | Descriptor: | (2S,4S)-5-carbamimidamido-4-hydroxy-2-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]pentanoic acid (non-preferred name), Aminotransferase class I/II-fold pyridoxal phosphate-dependent enzyme, MAGNESIUM ION | | Authors: | Chen, P.Y.-T, Moore, B.S. | | Deposit date: | 2022-12-10 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Mechanistic and Structural Insights into a Divergent PLP-Dependent l-Enduracididine Cyclase from a Toxic Cyanobacterium.
Acs Catalysis, 13, 2023
|
|
6OHI
 
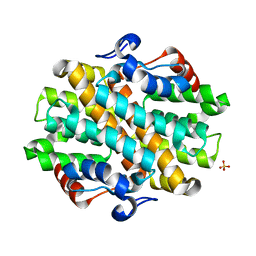 | | Crystal Structure of the Debrominase Bmp8 (Apo) | | Descriptor: | Debrominase Bmp8, SULFATE ION | | Authors: | Chekan, J.R, Moore, B.S. | | Deposit date: | 2019-04-05 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Bacterial Tetrabromopyrrole Debrominase Shares a Reductive Dehalogenation Strategy with Human Thyroid Deiodinase.
Biochemistry, 58, 2019
|
|
8CXL
 
 | | Structure of NapH3, a vanadium-dependent haloperoxidase homolog catalyzing the stereospecific alpha-hydroxyketone rearrangement reaction in napyradiomycin biosynthesis | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NapH3 | | Authors: | Chen, P.Y.-T, Chekan, J.R, Moore, B.S. | | Deposit date: | 2022-05-21 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural Basis of Stereospecific Vanadium-Dependent Haloperoxidase Family Enzymes in Napyradiomycin Biosynthesis.
Biochemistry, 61, 2022
|
|
7S5L
 
 | |
6OHJ
 
 | | Crystal Structure of the Debrominase Bmp8 C82A in Complex with 2,3,4-tribromopyrrole | | Descriptor: | 2,3,4-tribromo-1H-pyrrole, Debrominase Bmp8, SULFATE ION | | Authors: | Chekan, J.R, Moore, B.S. | | Deposit date: | 2019-04-05 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Bacterial Tetrabromopyrrole Debrominase Shares a Reductive Dehalogenation Strategy with Human Thyroid Deiodinase.
Biochemistry, 58, 2019
|
|
6NSD
 
 | | Tar14, tryptophan C-6 flavin-dependent halogenase (chlorinase) from taromycin biosynthesis | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, Tryptophan halogenase | | Authors: | Luhavaya, H, Chekan, J.R, Moore, B.S. | | Deposit date: | 2019-01-24 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Biosynthesis of l-4-Chlorokynurenine, an Antidepressant Prodrug and a Non-Proteinogenic Amino Acid Found in Lipopeptide Antibiotics.
Angew.Chem.Int.Ed.Engl., 58, 2019
|
|
7S2X
 
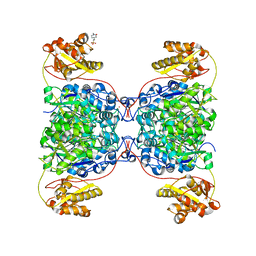 | | Structure of SalC, a gamma-lactam-beta-lactone bicyclase for salinosporamide biosynthesis | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, POTASSIUM ION, SalC | | Authors: | Chen, P.Y.-T, Trivella, D.B.B, Bauman, K.D, Moore, B.S. | | Deposit date: | 2021-09-04 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Enzymatic assembly of the salinosporamide gamma-lactam-beta-lactone anticancer warhead.
Nat.Chem.Biol., 18, 2022
|
|
5BUL
 
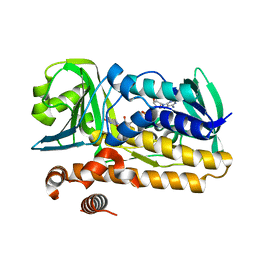 | | Structure of flavin-dependent brominase Bmp2 triple mutant Y302S F306V A345W | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, flavin-dependent halogenase triple mutant | | Authors: | Agarwal, V, Louie, G.V, Noel, J.P, Moore, B.S. | | Deposit date: | 2015-06-04 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9784 Å) | | Cite: | Biosynthesis of coral settlement cue tetrabromopyrrole in marine bacteria by a uniquely adapted brominase-thioesterase enzyme pair.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5BVA
 
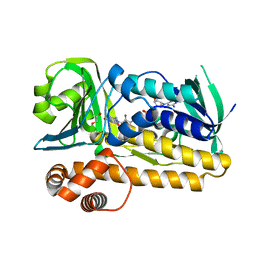 | | Structure of flavin-dependent brominase Bmp2 | | Descriptor: | 1,2-ETHANEDIOL, FLAVIN-ADENINE DINUCLEOTIDE, flavin-dependent halogenase | | Authors: | Agarwal, V, Louie, G.V, Noel, J.P, Moore, B.S. | | Deposit date: | 2015-06-04 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.873 Å) | | Cite: | Biosynthesis of coral settlement cue tetrabromopyrrole in marine bacteria by a uniquely adapted brominase-thioesterase enzyme pair.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6VKZ
 
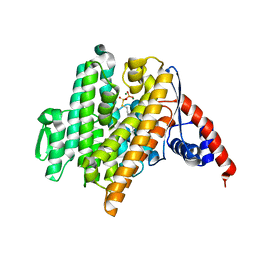 | |
6VL1
 
 | | Crystal Structure of the N-prenyltransferase DabA in Complex with NGG and Mg2+ | | Descriptor: | DabA, MAGNESIUM ION, N-[(2E)-3,7-dimethylocta-2,6-dien-1-yl]-L-glutamic acid | | Authors: | Chekan, J.R, Noel, J.P, Moore, B.S. | | Deposit date: | 2020-01-22 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Algal neurotoxin biosynthesis repurposes the terpene cyclase structural fold into anN-prenyltransferase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VL0
 
 | | Crystal Structure of the N-prenyltransferase DabA in Complex with GSPP and Mn2+ | | Descriptor: | DabA, GERANYL S-THIOLODIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Chekan, J.R, Noel, J.P, Moore, B.S. | | Deposit date: | 2020-01-22 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Algal neurotoxin biosynthesis repurposes the terpene cyclase structural fold into anN-prenyltransferase.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5BUK
 
 | | Structure of flavin-dependent chlorinase Mpy16 | | Descriptor: | FADH2-dependent halogenase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Agarwal, V, Louie, G.V, Noel, J.P, Moore, B.S. | | Deposit date: | 2015-06-03 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Biosynthesis of coral settlement cue tetrabromopyrrole in marine bacteria by a uniquely adapted brominase-thioesterase enzyme pair.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3W35
 
 | | Crystal structure of apo-type bacterial Vanadium-dependent chloroperoxidase | | Descriptor: | NapH1 | | Authors: | Liscombe, D.K, Miyanaga, A, Fielding, E, Bernhardt, P, Li, A, Winter, J.M, Gilson, M.K, Noel, J.P, Moore, B.S. | | Deposit date: | 2012-12-11 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Stereospecific Vanadium-Dependent Haloperoxidase Family Enzymes in Napyradiomycin Biosynthesis.
Biochemistry, 2022
|
|
3W36
 
 | | Crystal structure of holo-type bacterial Vanadium-dependent chloroperoxidase | | Descriptor: | NapH1, VANADATE ION | | Authors: | Liscombe, D.K, Miyanaga, A, Fielding, E, Bernhardt, P, Li, A, Winter, J.M, Gilson, M.K, Noel, J.P, Moore, B.S. | | Deposit date: | 2012-12-11 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis of Stereospecific Vanadium-Dependent Haloperoxidase Family Enzymes in Napyradiomycin Biosynthesis.
Biochemistry, 2022
|
|
3W8W
 
 | | The crystal structure of EncM | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Putative FAD-dependent oxygenase EncM | | Authors: | Teufel, R, Miyanaga, A, Stull, F, Michaudel, Q, Louie, G, Noel, J.P, Baran, P.S, Palfey, B, Moore, B.S. | | Deposit date: | 2013-03-22 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Flavin-mediated dual oxidation controls an enzymatic Favorskii-type rearrangement.
Nature, 503, 2013
|
|
3W8Z
 
 | | The complex structure of EncM with hydroxytetraketide | | Descriptor: | (7S)-7-hydroxy-1-phenyloctane-1,3,5-trione, FLAVIN-ADENINE DINUCLEOTIDE, Putative FAD-dependent oxygenase EncM | | Authors: | Teufel, R, Miyanaga, A, Stull, F, Michaudel, Q, Louie, G, Noel, J.P, Baran, P.S, Palfey, B, Moore, B.S. | | Deposit date: | 2013-03-22 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Flavin-mediated dual oxidation controls an enzymatic Favorskii-type rearrangement.
Nature, 503, 2013
|
|
3W8X
 
 | | The complex structure of EncM with trifluorotriketide | | Descriptor: | 6,6,6-trifluoro-1-phenylhexane-1,3,5-trione, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Teufel, R, Miyanaga, A, Stull, F, Michaudel, Q, Louie, G, Noel, J.P, Baran, P.S, Palfey, B, Moore, B.S. | | Deposit date: | 2013-03-22 | | Release date: | 2013-10-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Flavin-mediated dual oxidation controls an enzymatic Favorskii-type rearrangement.
Nature, 503, 2013
|
|
2NYF
 
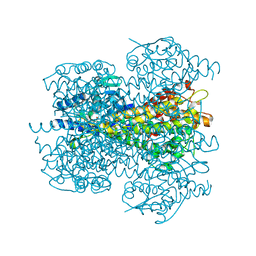 | | Crystal structure of phenylalanine ammonia-lyase from Nostoc punctiforme | | Descriptor: | Nostoc punctiforme phenylalanine ammonia lyase | | Authors: | Louie, G.V, Moffitt, M.C, Bowman, M.E, Pence, J, Noel, J.P, Moore, B.S. | | Deposit date: | 2006-11-20 | | Release date: | 2007-02-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of Two Cyanobacterial Phenylalanine Ammonia Lyases: Kinetic and Structural Characterization.
Biochemistry, 46, 2007
|
|
2NYN
 
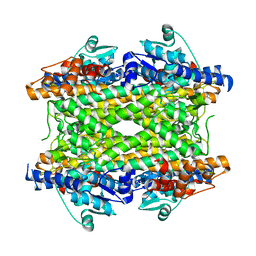 | | Crystal structure of phenylalanine ammonia-lyase from Anabaena variabilis | | Descriptor: | Phenylalanine/histidine ammonia-lyase | | Authors: | Louie, G.V, Moffitt, M.C, Bowman, M.E, Pence, J, Noel, J.P, Moore, B.S. | | Deposit date: | 2006-11-21 | | Release date: | 2007-02-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Two Cyanobacterial Phenylalanine Ammonia Lyases: Kinetic and Structural Characterization.
Biochemistry, 46, 2007
|
|
2H84
 
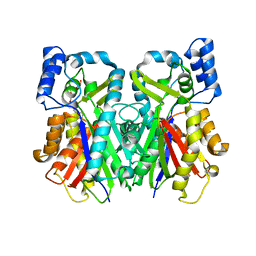 | | Crystal Structure of the C-terminal Type III Polyketide Synthase (PKS III) Domain of 'Steely1' (a Type I/III PKS Hybrid from Dictyostelium) | | Descriptor: | HEXAETHYLENE GLYCOL, Steely1 | | Authors: | Austin, M.B, Saito, T, Bowman, M.E, Haydock, S, Kato, A, Moore, B.S, Kay, R.R, Noel, J.P. | | Deposit date: | 2006-06-06 | | Release date: | 2006-08-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Biosynthesis of Dictyostelium discoideum differentiation-inducing factor by a hybrid type I fatty acid-type III polyketide synthase.
Nat.Chem.Biol., 2, 2006
|
|
