7Q41
 
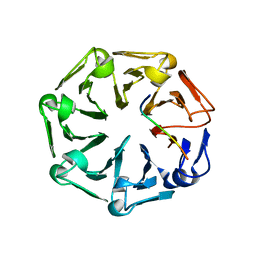 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of ubiquitin-protein ligase E3A (E6AP) | | 分子名称: | CITRIC ACID, E3 ubiquitin-protein ligase HERC2, Ubiquitin-protein ligase E3A (E6AP) peptide | | 著者 | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | 登録日 | 2021-10-29 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (3.01478052 Å) | | 主引用文献 | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of ubiquitin-protein ligase E3A (E6AP)
To Be Published
|
|
7Q45
 
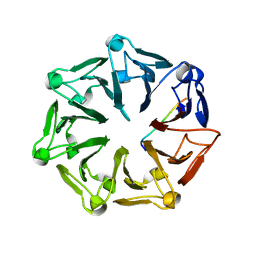 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of Myelin transcription factor 1 | | 分子名称: | CITRIC ACID, E3 ubiquitin-protein ligase HERC2, Myelin transcription factor 1 | | 著者 | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | 登録日 | 2021-10-29 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.09999585 Å) | | 主引用文献 | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of Myelin transcription factor 1
To Be Published
|
|
7Q42
 
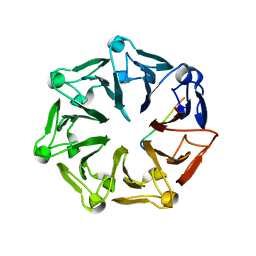 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of chromatin reader BAZ2B | | 分子名称: | Bromodomain adjacent to zinc finger domain protein 2B, CITRIC ACID, E3 ubiquitin-protein ligase HERC2 | | 著者 | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | 登録日 | 2021-10-29 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.95002484 Å) | | 主引用文献 | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of chromatin reader BAZ2B
To Be Published
|
|
7Q40
 
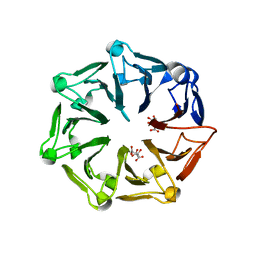 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 | | 分子名称: | CITRIC ACID, E3 ubiquitin-protein ligase HERC2 | | 著者 | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | 登録日 | 2021-10-29 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.35002232 Å) | | 主引用文献 | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2
To Be Published
|
|
7Q44
 
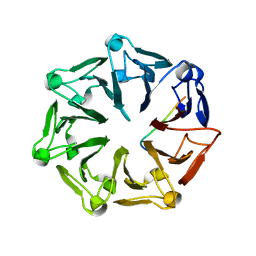 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of deubiquitinase USP35 | | 分子名称: | CITRIC ACID, Deubiquitinase USP35 peptide, E3 ubiquitin-protein ligase HERC2 | | 著者 | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | 登録日 | 2021-10-29 | | 公開日 | 2022-11-16 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.20007777 Å) | | 主引用文献 | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of deubiquitinase USP35
To Be Published
|
|
6SIV
 
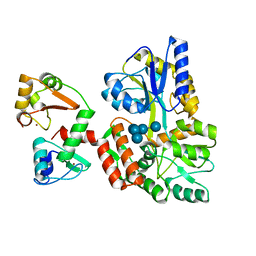 | | Structure of HPV16 E6 oncoprotein in complex with mutant IRF3 LxxLL motif | | 分子名称: | Maltose/maltodextrin-binding periplasmic protein,Interferon regulatory factor 3, Protein E6, ZINC ION, ... | | 著者 | Suarez, I.P, Cousido-Siah, A, Bonhoure, A, Mitschler, A, Podjarny, A, Trave, G. | | 登録日 | 2019-08-12 | | 公開日 | 2019-08-21 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (1.752 Å) | | 主引用文献 | Deciphering the molecular and structural interaction between IRF3 and HPV16 E6
To be published
|
|
2PEV
 
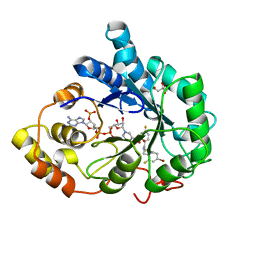 | | Complex of Aldose Reductase with NADP+ and simaltaneously bound competetive inhibitors Fidarestat and IDD594. Concentration of Fidarestat in soaking solution exceeds concentration of IDD594. | | 分子名称: | (2S,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose reductase, CHLORIDE ION, ... | | 著者 | Petrova, T, Hazemann, I, Cousido, A, Mitschler, A, Ginell, S, Joachimiak, A, Podjarny, A. | | 登録日 | 2007-04-03 | | 公開日 | 2007-04-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (0.9 Å) | | 主引用文献 | Crystal packing modifies ligand binding affinity: The case of aldose reductase.
Proteins, 80, 2012
|
|
2PFH
 
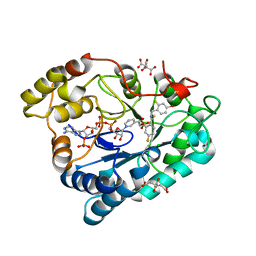 | | Complex of Aldose Reductase with NADP+ and simaltaneously bound competetive inhibitors Fidarestat and IDD594. Concentration of Fidarestat in soaking solution is less than concentration of IDD594. | | 分子名称: | (2S,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose reductase, CHLORIDE ION, ... | | 著者 | Petrova, T, Hazemann, I, Cousido, A, Mitschler, A, Ginell, S, Joachimiak, A, Podjarny, A. | | 登録日 | 2007-04-05 | | 公開日 | 2007-04-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Crystal packing modifies ligand binding affinity: The case of aldose reductase.
Proteins, 80, 2012
|
|
2PF8
 
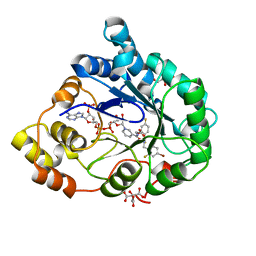 | | Complex of Aldose Reductase with NADP+ and simaltaneously bound competetive inhibitors Fidarestat and IDD594. Concentration of Fidarestat in soaking solution is equal to concentration of IDD594. | | 分子名称: | (2S,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose reductase, CHLORIDE ION, ... | | 著者 | Petrova, T, Hazemann, I, Cousido, A, Mitschler, A, Ginell, S, Joachimiak, A, Podjarny, A. | | 登録日 | 2007-04-04 | | 公開日 | 2007-04-17 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (0.85 Å) | | 主引用文献 | Crystal packing modifies ligand binding affinity: The case of aldose reductase.
Proteins, 80, 2012
|
|
1US0
 
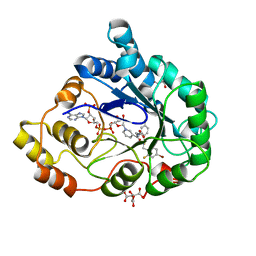 | | Human Aldose Reductase in complex with NADP+ and the inhibitor IDD594 at 0.66 Angstrom | | 分子名称: | ALDOSE REDUCTASE, CITRIC ACID, IDD594, ... | | 著者 | Howard, E.I, Sanishvili, R, Cachau, R.E, Mitschler, A, Chevrier, B, Barth, P, Lamour, V, Van Zandt, M, Sibley, E, Bon, C, Moras, D, Schneider, T.R, Joachimiak, A, Podjarny, A. | | 登録日 | 2003-11-16 | | 公開日 | 2004-05-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (0.66 Å) | | 主引用文献 | Ultrahigh Resolution Drug Design I: Details of Interactions in Human Aldose Reductase-Inhibitor Complex at 0.66 A.
Proteins, 55, 2004
|
|
2QXW
 
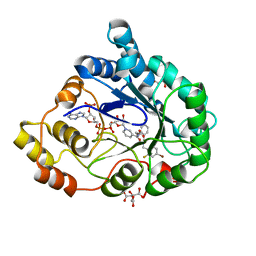 | | Perdeuterated alr2 in complex with idd594 | | 分子名称: | Aldose reductase, CITRIC ACID, IDD594, ... | | 著者 | Blakeley, M.P, Ruiz, F, Cachau, R, Hazemann, I, Meilleur, F, Mitschler, A, Ginell, S, Afonine, P, Ventura, O, Cousido-Siah, A, Joachimiak, A, Myles, D, Podjarny, A. | | 登録日 | 2007-08-13 | | 公開日 | 2008-01-22 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (0.8 Å) | | 主引用文献 | Quantum model of catalysis based on a mobile proton revealed by subatomic x-ray and neutron diffraction studies of h-aldose reductase.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1T41
 
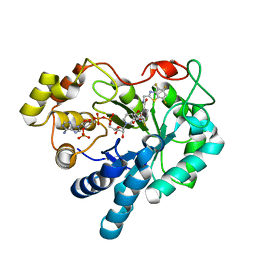 | | Crystal structure of human aldose reductase complexed with NADP and IDD552 | | 分子名称: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [5-FLUORO-2-({[(4,5,7-TRIFLUORO-1,3-BENZOTHIAZOL-2-YL)METHYL]AMINO}CARBONYL)PHENOXY]ACETIC ACID | | 著者 | Ruiz, F, Hazemann, I, Mitschler, A, Chevrier, B, Schneider, T, Joachimiak, A, Karplus, M, Podjarny, A. | | 登録日 | 2004-04-28 | | 公開日 | 2004-08-03 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | The crystallographic structure of the aldose reductase-IDD552 complex shows direct proton donation from tyrosine 48.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
5NRF
 
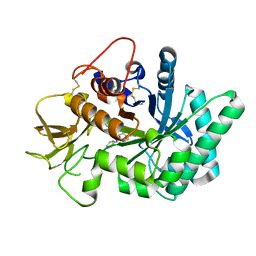 | | Crystal structure of human chitotriosidase-1 (hCHIT) catalytic domain in complex with compound 7i | | 分子名称: | 1-(3-azanyl-1~{H}-1,2,4-triazol-5-yl)-~{N}-[2-(4-chlorophenyl)ethyl]-~{N}-(phenylmethyl)piperidin-4-amine, Chitotriosidase-1, GLYCEROL | | 著者 | Mazur, M, Olczak, J, Olejniczak, S, Koralewski, R, Czestkowski, W, Jedrzejczak, A, Golab, J, Dzwonek, K, Dymek, B, Sklepkiewicz, P, Zagozdzon, A, Noonan, T, Mahboubi, K, Conway, B, Sheeler, R, Beckett, P, Hungerford, W.M, Podjarny, A, Mitschler, A, Cousido-Siah, A, Fadel, F, Golebiowski, A. | | 登録日 | 2017-04-22 | | 公開日 | 2018-03-28 | | 実験手法 | X-RAY DIFFRACTION (1.447 Å) | | 主引用文献 | Targeting Acidic Mammalian chitinase Is Effective in Animal Model of Asthma.
J. Med. Chem., 61, 2018
|
|
5M2F
 
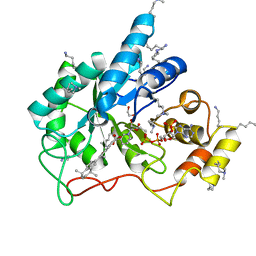 | | Crystal structure of human AKR1B10 complexed with NADP+ and the synthetic retinoid UVI2008 | | 分子名称: | 1,2-ETHANEDIOL, 3-bromo-4-[(1E)-2-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydronaphthalen-2-yl)prop-1-en-1-yl]benzoic acid, Aldo-keto reductase family 1 member B10, ... | | 著者 | Ruiz, F.X, Cousido-Siah, A, Mitschler, A, Porte, S, Alvarez, S, Dominguez, M, Alvarez, R, de Lera, A.R, Pares, X, Farres, J, Podjarny, A. | | 登録日 | 2016-10-12 | | 公開日 | 2017-02-15 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.503 Å) | | 主引用文献 | Structural basis for the inhibition of AKR1B10 by the C3 brominated TTNPB derivative UVI2008.
Chem. Biol. Interact., 276, 2017
|
|
1T40
 
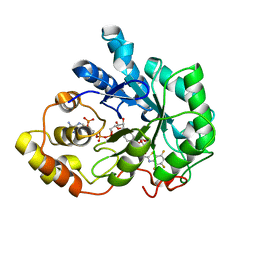 | | Crystal structure of human aldose reductase complexed with NADP and IDD552 at ph 5 | | 分子名称: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [5-FLUORO-2-({[(4,5,7-TRIFLUORO-1,3-BENZOTHIAZOL-2-YL)METHYL]AMINO}CARBONYL)PHENOXY]ACETIC ACID | | 著者 | Ruiz, F, Hazemann, I, Mitschler, A, Chevrier, B, Schneider, T, Joachimiak, A, Karplus, M, Podjarny, A. | | 登録日 | 2004-04-28 | | 公開日 | 2004-08-03 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | The crystallographic structure of the aldose reductase-IDD552 complex shows direct proton donation from tyrosine 48.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3GHU
 
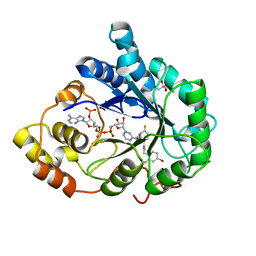 | | Human aldose reductase in complex with NADP+ and the inhibitor IDD594. Investigation of global effects of radiation damage on protein structure. Forth stage of radiation damage. | | 分子名称: | Aldose reductase, CITRIC ACID, IDD594, ... | | 著者 | Petrova, T, Ginell, S, Hazemann, I, Mitschler, A, Podjarny, A, Joachimiak, A. | | 登録日 | 2009-03-04 | | 公開日 | 2009-03-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | X-ray-radiation-induced cooperative atomic movements in protein.
J.Mol.Biol., 387, 2009
|
|
3GHS
 
 | | Human aldose reductase in complex with NADP+ and the inhibitor IDD594. Investigation of global effects of radiation damage on protein structure. Second stage of radiation damage. | | 分子名称: | Aldose reductase, CITRIC ACID, IDD594, ... | | 著者 | Petrova, T, Ginell, S, Hazemann, I, Mitschler, A, Podjarny, A, Joachimiak, A. | | 登録日 | 2009-03-04 | | 公開日 | 2009-03-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | X-ray-radiation-induced cooperative atomic movements in protein.
J.Mol.Biol., 387, 2009
|
|
3GHT
 
 | | Human aldose reductase in complex with NADP+ and the inhibitor IDD594. Investigation of global effects of radiation damage on protein structure. Third stage of radiation damage. | | 分子名称: | Aldose reductase, CITRIC ACID, IDD594, ... | | 著者 | Petrova, T, Ginell, S, Hazemann, I, Mitschler, A, Podjarny, A, Joachimiak, A. | | 登録日 | 2009-03-04 | | 公開日 | 2009-03-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | X-ray-radiation-induced cooperative atomic movements in protein.
J.Mol.Biol., 387, 2009
|
|
3GHR
 
 | | Human aldose reductase in complex with NADP+ and the inhibitor IDD594. Investigation of global effects of radiation damage on protein structure. First stage of radiation damage | | 分子名称: | Aldose reductase, CITRIC ACID, IDD594, ... | | 著者 | Petrova, T, Ginell, S, Hazemann, I, Mitschler, A, Podjarny, A, Joachimiak, A. | | 登録日 | 2009-03-04 | | 公開日 | 2009-03-24 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | X-ray-radiation-induced cooperative atomic movements in protein.
J.Mol.Biol., 387, 2009
|
|
5LIK
 
 | | Crystal structure of human AKR1B10 complexed with NADP+ and the inhibitor MK181 | | 分子名称: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Fanfrlik, J, Kamlar, M, Vesely, J, Hobza, P, Podjarny, A. | | 登録日 | 2016-07-14 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | IDD388 Polyhalogenated Derivatives as Probes for an Improved Structure-Based Selectivity of AKR1B10 Inhibitors.
Acs Chem.Biol., 11, 2016
|
|
5LIY
 
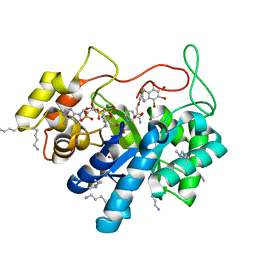 | | Crystal structure of human AKR1B10 complexed with NADP+ and the inhibitor MK204 | | 分子名称: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Fanfrlik, J, Kamlar, M, Vesely, J, Hobza, P, Podjarny, A. | | 登録日 | 2016-07-15 | | 公開日 | 2016-10-12 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | IDD388 Polyhalogenated Derivatives as Probes for an Improved Structure-Based Selectivity of AKR1B10 Inhibitors.
Acs Chem.Biol., 11, 2016
|
|
5LIU
 
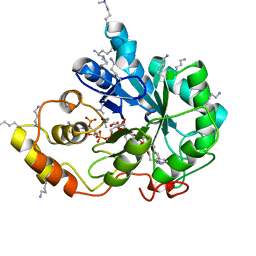 | | Crystal structure of human AKR1B10 complexed with NADP+ and the inhibitor IDD388 | | 分子名称: | (2-{[(4-BROMO-2-FLUOROBENZYL)AMINO]CARBONYL}-5-CHLOROPHENOXY)ACETIC ACID, 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, ... | | 著者 | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Fanfrlik, J, Kamlar, M, Vesely, J, Hobza, P, Podjarny, A. | | 登録日 | 2016-07-15 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | IDD388 Polyhalogenated Derivatives as Probes for an Improved Structure-Based Selectivity of AKR1B10 Inhibitors.
Acs Chem.Biol., 11, 2016
|
|
5LIW
 
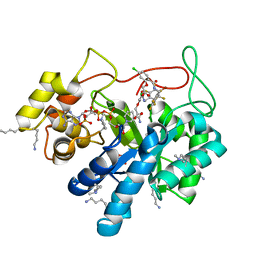 | | Crystal structure of human AKR1B10 complexed with NADP+ and the inhibitor MK319 | | 分子名称: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Fanfrlik, J, Kamlar, M, Vesely, J, Hobza, P, Podjarny, A. | | 登録日 | 2016-07-15 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | IDD388 Polyhalogenated Derivatives as Probes for an Improved Structure-Based Selectivity of AKR1B10 Inhibitors.
Acs Chem.Biol., 11, 2016
|
|
6SLM
 
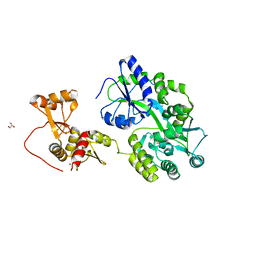 | | Crystal structure of full-length HPV31 E6 oncoprotein in complex with LXXLL peptide of ubiquitin ligase E6AP | | 分子名称: | GLYCEROL, Maltose/maltodextrin-binding periplasmic protein,Protein E6,Ubiquitin-protein ligase E3A, ZINC ION, ... | | 著者 | Conrady, M, Gogl, G, Cousido-Siah, A, Mitschler, A, Trave, G, Simon, C. | | 登録日 | 2019-08-20 | | 公開日 | 2020-09-09 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of High-Risk Papillomavirus 31 E6 Oncogenic Protein and Characterization of E6/E6AP/p53 Complex Formation.
J.Virol., 95, 2020
|
|
5LIX
 
 | | Crystal structure of human AKR1B10 complexed with NADP+ and the inhibitor MK184 | | 分子名称: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Fanfrlik, J, Kamlar, M, Vesely, J, Hobza, P, Podjarny, A. | | 登録日 | 2016-07-15 | | 公開日 | 2016-07-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | IDD388 Polyhalogenated Derivatives as Probes for an Improved Structure-Based Selectivity of AKR1B10 Inhibitors.
Acs Chem.Biol., 11, 2016
|
|
