5GO1
 
 | | Structural, Functional characterization and discovery of novel inhibitors of Leishmania amazonensis Nucleoside Diphosphatase Kinase (NDK) | | Descriptor: | Nucleoside diphosphate kinase | | Authors: | Mishra, A.K, Agnihotri, P, Singh, S.P, Pratap, J.V. | | Deposit date: | 2016-07-26 | | Release date: | 2017-07-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of novel inhibitors for Leishmania nucleoside diphosphatase kinase (NDK) based on its structural and functional characterization.
J. Comput. Aided Mol. Des., 31, 2017
|
|
8SO3
 
 | | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte activation gene 3 protein, favezelimab Fab heavy chain, ... | | Authors: | Mishra, A.K, Shahid, S, Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2023-04-28 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 determined using a bivalent Fab as fiducial marker.
Structure, 31, 2023
|
|
8SR0
 
 | | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 local refined | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte activation gene 3 protein, favezelimab Fab heavy chain, ... | | Authors: | Mishra, A.K, Shahid, S, Karade, S.S, Mariuzza, R.A. | | Deposit date: | 2023-05-05 | | Release date: | 2023-09-06 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 determined using a bivalent Fab as fiducial marker.
Structure, 31, 2023
|
|
6VKF
 
 | | CCHFV GP38 (IbAr10200) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GP38 | | Authors: | Mishra, A.K, McLellan, J.S. | | Deposit date: | 2020-01-20 | | Release date: | 2020-02-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.524 Å) | | Cite: | Structure and Characterization of Crimean-Congo Hemorrhagic Fever Virus GP38.
J.Virol., 94, 2020
|
|
3MJH
 
 | | Crystal Structure of Human Rab5A in complex with the C2H2 Zinc Finger of EEA1 | | Descriptor: | Early endosome antigen 1, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Mishra, A.K, Eathiraj, S, Lambright, D.G. | | Deposit date: | 2010-04-12 | | Release date: | 2010-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural basis for Rab GTPase recognition and endosome tethering by the C2H2 zinc finger of Early Endosomal Autoantigen 1 (EEA1).
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2QKD
 
 | | Crystal structure of tandem ZPR1 domains | | Descriptor: | FORMIC ACID, ZINC ION, Zinc finger protein ZPR1 | | Authors: | Mishra, A.K. | | Deposit date: | 2007-07-10 | | Release date: | 2007-08-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insights into the interaction of the evolutionarily conserved ZPR1 domain tandem with eukaryotic EF1A, receptors, and SMN complexes.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4IRU
 
 | | Crystal Structure of lepB GAP core in a transition state mimetic complex with Rab1A and ALF3 | | Descriptor: | ACETATE ION, ALUMINUM FLUORIDE, GLYCEROL, ... | | Authors: | Mishra, A.K, Delcampo, C.M, Collins, R.E, Roy, C.R, Lambright, D.G. | | Deposit date: | 2013-01-15 | | Release date: | 2013-07-10 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Legionella pneumophila GTPase Activating Protein LepB Accelerates Rab1 Deactivation by a Non-canonical Hydrolytic Mechanism.
J.Biol.Chem., 288, 2013
|
|
8FWH
 
 | | Crystal structure of bivalent antibody Fab fragment of Anti-human LAG3 (22D2) | | Descriptor: | 1,2-ETHANEDIOL, Anti-human LAG3 (22D2) heavy chain, Anti-human LAG3 (22D2) light chain | | Authors: | Mishra, A.K, Agnihotri, P, Mariuzza, R.A. | | Deposit date: | 2023-01-22 | | Release date: | 2023-09-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.833 Å) | | Cite: | CryoEM structure of a therapeutic antibody (favezelimab) bound to human LAG3 determined using a bivalent Fab as fiducial marker.
Structure, 31, 2023
|
|
7KX4
 
 | | Anti-CCHFV ADI-36121 Fab | | Descriptor: | ADI-36121 Fab heavy chain, ADI-36121 Fab light chain | | Authors: | Mishra, A.K, McLellan, J.S. | | Deposit date: | 2020-12-03 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of synergistic neutralization of Crimean-Congo hemorrhagic fever virus by human antibodies.
Science, 375, 2022
|
|
7L7R
 
 | |
6JCH
 
 | | Crystal structure of SpaE basal pilin from Lactobacillus rhamnosus GG - Orthorhombic form | | Descriptor: | Pilus assembly protein, SODIUM ION | | Authors: | Megta, A.K, Mishra, A.K, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2019-01-28 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.536 Å) | | Cite: | Crystal structure of basal pilin SpaE reveals the molecular basis of its incorporation in the lactobacillar SpaFED pilus.
J.Struct.Biol., 207, 2019
|
|
6JBV
 
 | | Crystal structure of SpaE basal pilin from Lactobacillus rhamnosus GG - Selenium derivative | | Descriptor: | Pilus assembly protein, SODIUM ION | | Authors: | Megta, A.K, Mishra, A.K, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2019-01-26 | | Release date: | 2019-06-26 | | Last modified: | 2021-09-15 | | Method: | X-RAY DIFFRACTION (1.712 Å) | | Cite: | Crystal structure of basal pilin SpaE reveals the molecular basis of its incorporation in the lactobacillar SpaFED pilus.
J.Struct.Biol., 207, 2019
|
|
6JK7
 
 | | Crystal structure of SpaE basal pilin from Lactobacillus rhamnosus GG - Trigonal form | | Descriptor: | Pilus assembly protein | | Authors: | Megta, A.K, Mishra, A.K, Palva, A, von Ossowski, I, Krishnan, V. | | Deposit date: | 2019-02-27 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.204 Å) | | Cite: | Crystal structure of basal pilin SpaE reveals the molecular basis of its incorporation in the lactobacillar SpaFED pilus.
J.Struct.Biol., 207, 2019
|
|
5X00
 
 | |
8VVL
 
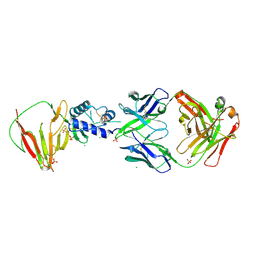 | | CCHFV GP38 bound to c13G8 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Hjorth, C.K, Mishra, A.K, McLellan, J.S. | | Deposit date: | 2024-01-31 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crimean-Congo hemorrhagic fever survivors elicit protective non-neutralizing antibodies that target 11 overlapping regions on glycoprotein GP38.
Cell Rep, 43, 2024
|
|
6WJU
 
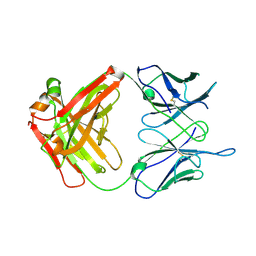 | |
6WK4
 
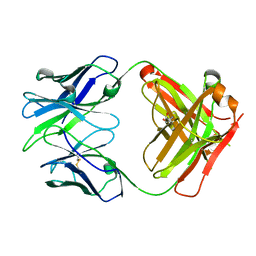 | |
6WKL
 
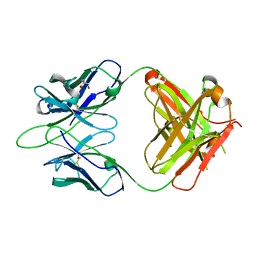 | |
6WKM
 
 | |
4MXP
 
 | | Structural Basis for PI(4)P-Specific Membrane Recruitment of the Legionella pneumophila Effector DrrA/SidM | | Descriptor: | (2R)-3-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}propane-1,2-diyl dibutanoate, Defects in Rab1 recruitment protein A, SODIUM ION | | Authors: | Del Campo, C.M, Mishra, A.K, Wang, Y.H, Roy, C.R, Janmey, P.A, Lambright, D.G. | | Deposit date: | 2013-09-26 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Basis for PI(4)P-Specific Membrane Recruitment of the Legionella pneumophila Effector DrrA/SidM.
Structure, 22, 2014
|
|
7LYW
 
 | |
8E1B
 
 | |
8F6U
 
 | |
