2FK4
 
 | |
2HFR
 
 | | solution structure of antimicrobial peptide Fowlicidin 3 | | Descriptor: | Fowlicidin-3 | | Authors: | Bommineni, Y.R, Dai, H, Gong, Y, Prakash, O, Zhang, G. | | Deposit date: | 2006-06-26 | | Release date: | 2007-04-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Fowlicidin-3 is an alpha-helical cationic host defense peptide with potent antibacterial and lipopolysaccharide-neutralizing activities.
Febs J., 274, 2007
|
|
2KRE
 
 | |
6KX0
 
 | | Crystal structure of SN-101 mAb non-liganded form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Fab Fragment-SN-101-Heavy chain, Fab Fragment-SN-101-Light chain | | Authors: | Wakui, H, Tanaka, Y, Kato, K, Ose, T, Matsumoto, I, Min, Y, Tachibana, T, Nishimura, S.-I. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | A straightforward approach to antibodies recognising cancer specific glycopeptidic neoepitopes
Chem Sci, 11, 2020
|
|
6KX1
 
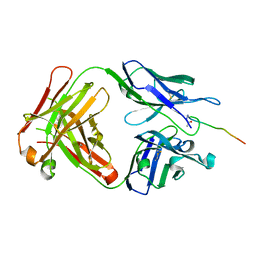 | | Crystal structure of SN-101 mAb in complex with MUC1 glycopeptide | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Fab Fragment-SN-101-Heavy chain, Fab Fragment-SN-101-Light chain, ... | | Authors: | Wakui, H, Tanaka, Y, Kato, K, Ose, T, Matsumoto, I, Min, Y, Tachibana, T, Nishimura, S.-I. | | Deposit date: | 2019-09-09 | | Release date: | 2020-07-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | A straightforward approach to antibodies recognising cancer specific glycopeptidic neoepitopes
Chem Sci, 11, 2020
|
|
6KNE
 
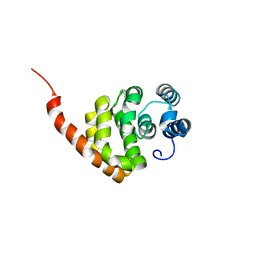 | |
6KND
 
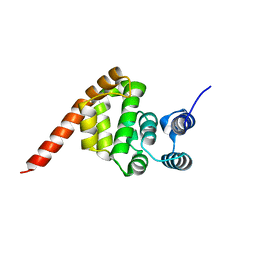 | |
2E6U
 
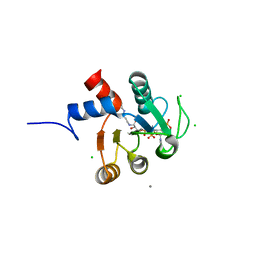 | | Crystal structure of hypothetical protein PH1109 from Pyrococcus horikoshii | | Descriptor: | CALCIUM ION, CHLORIDE ION, COENZYME A, ... | | Authors: | Kitago, Y, Min, Y, Watanabe, N, Tanaka, I. | | Deposit date: | 2007-01-03 | | Release date: | 2007-01-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure determination of a novel protein by sulfur SAD using chromium radiation in combination with a new crystal-mounting method
ACTA CRYSTALLOGR.,SECT.D, 61, 2005
|
|
3ZJB
 
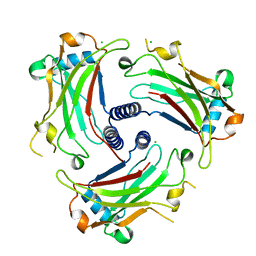 | | The structure of the TRAF domain of human TRAF4 | | Descriptor: | CHLORIDE ION, TNF RECEPTOR-ASSOCIATED FACTOR 4 | | Authors: | McEwen, A.G, Poussin-Courmontagne, P, Rousseau, A, Rogna, D, Nomine, Y, Rio, M.-C, Tomasetto, C, Alpy, F. | | Deposit date: | 2013-01-17 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Traf4 is a Novel Phosphoinositide-Binding Protein Modulating Tight Junctions and Favoring Cell Migration.
Plos Biol., 11, 2013
|
|
8I4O
 
 | | Design of a split green fluorescent protein for sensing and tracking an beta-amyloid | | Descriptor: | Beta-amyloid, Split Green flourescent protein | | Authors: | Taegeun, Y, Jinsu, L, Jungmin, Y, Jungmin, C, Wondo, H, Song, J.J, Haksung, K. | | Deposit date: | 2023-01-20 | | Release date: | 2023-11-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Engineering of a Fluorescent Protein for a Sensing of an Intrinsically Disordered Protein through Transition in the Chromophore State.
Jacs Au, 3, 2023
|
|
7BOS
 
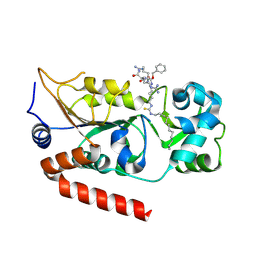 | | Human SIRT2 in complex with myristoyl thiourea inhibitor, No.13 | | Descriptor: | Myristoyl thiourea inhibitor, No.13, N-dodecylmethanethioamide, ... | | Authors: | Kudo, N, Olsen, C.A, Minoru, Y. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism-based inhibitors of SIRT2: structure-activity relationship, X-ray structures, target engagement, regulation of alpha-tubulin acetylation and inhibition of breast cancer cell migration.
Rsc Chem Biol, 2, 2021
|
|
7BOT
 
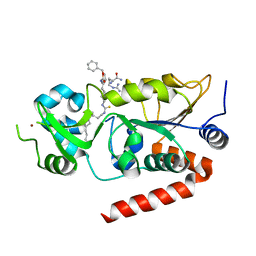 | | Human SIRT2 in complex with myristoyl thiourea inhibitor, No.23 | | Descriptor: | N-dodecylmethanethioamide, NAD-dependent protein deacetylase sirtuin-2, ZINC ION, ... | | Authors: | Kudo, N, Olsen, C.A, Minoru, Y. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism-based inhibitors of SIRT2: structure-activity relationship, X-ray structures, target engagement, regulation of alpha-tubulin acetylation and inhibition of breast cancer cell migration.
Rsc Chem Biol, 2, 2021
|
|
7TWU
 
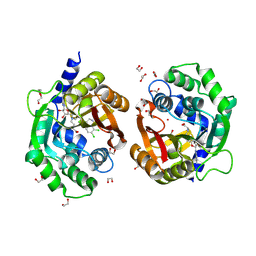 | | Crystal structure of human phenylethanolamine N-methyltransferase (PNMT) in complex with (2S)-2-amino-4-(((5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)(4-(7,8-dichloro-1,2,3,4-tetrahydroisoquinolin-4-yl)butyl)amino)butanoic acid and AdoHcy (SAH) | | Descriptor: | 1,2-ETHANEDIOL, 5'-([(3S)-3-amino-3-carboxypropyl]{4-[(4R)-7,8-dichloro-1,2,3,4-tetrahydroisoquinolin-4-yl]butyl}amino)-5'-deoxyadenosine, CADMIUM ION, ... | | Authors: | Harijan, R.K, Mahmoodi, N, Minnow, Y.V.T, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2022-02-07 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cell-Effective Transition-State Analogue of Phenylethanolamine N -Methyltransferase.
Biochemistry, 62, 2023
|
|
7TX2
 
 | | Crystal structure of human phenylethanolamine N-methyltransferase (PNMT) in complex with (2S)-2-amino-4-(((5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl)methyl)(4-(7,8-dichloro-1,2,3,4-tetrahydroisoquinolin-4-yl)butyl)amino)butanoic acid | | Descriptor: | 1,2-ETHANEDIOL, 5'-([(3S)-3-amino-3-carboxypropyl]{4-[(4R)-7,8-dichloro-1,2,3,4-tetrahydroisoquinolin-4-yl]butyl}amino)-5'-deoxyadenosine, Phenylethanolamine N-methyltransferase | | Authors: | Harijan, R.K, Mahmoodi, N, Minnow, Y.V.T, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2022-02-07 | | Release date: | 2023-02-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Cell-Effective Transition-State Analogue of Phenylethanolamine N -Methyltransferase.
Biochemistry, 62, 2023
|
|
1IO5
 
 | | HYDROGEN AND HYDRATION OF HEN EGG-WHITE LYSOZYME DETERMINED BY NEUTRON DIFFRACTION | | Descriptor: | LYSOZYME C | | Authors: | Niimura, N, Minezaki, Y, Nonaka, T, Castagna, J.C, Cipriani, F, Hoeghoej, P, Lehmann, M.S, Wilkinson, C. | | Deposit date: | 2001-01-14 | | Release date: | 2001-02-07 | | Last modified: | 2024-10-16 | | Method: | NEUTRON DIFFRACTION (2 Å) | | Cite: | Neutron Laue diffractometry with an imaging plate provides an effective data collection regime for neutron protein crystallography.
Nat.Struct.Biol., 4, 1997
|
|
7TUX
 
 | | Crystal Structure of Plasmodium falciparum Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferase in complex with [(3S)-4-Hydroxy-3-[({2-amino-4-hydroxy-5H-pyrrolo[3,2-d]pyrimidin-7-yl}methyl)amino]butyl]phosphonic acid | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Hypoxanthine-guanine-xanthine phosphoribosyltransferase, ... | | Authors: | Harijan, R.K, Minnow, Y.V.T, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2022-02-03 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Inhibition and Mechanism of Plasmodium falciparum Hypoxanthine-Guanine-Xanthine Phosphoribosyltransferase.
Acs Chem.Biol., 17, 2022
|
|
2EIX
 
 | | The Structure of Physarum polycephalum cytochrome b5 reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, IODIDE ION, ... | | Authors: | Kim, S.W, Suga, M, Ogasahara, K, Ikegami, T, Minami, Y, Yubisui, T, Tsukihara, T. | | Deposit date: | 2007-03-14 | | Release date: | 2007-04-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structure of Physarum polycephalum cytochrome b5 reductase at 1.56 A resolution.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
2B02
 
 | | Crystal Structure of ARNT PAS-B Domain | | Descriptor: | Aryl hydrocarbon receptor nuclear translocator | | Authors: | Lee, J, Botuyan, M.V, Nomine, Y, Ohh, M, Thompson, J.R, Mer, G. | | Deposit date: | 2005-09-12 | | Release date: | 2006-10-24 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure and Binding Properties of ARNT PAS-B Heterodimerization Domain
To be Published
|
|
2YZQ
 
 | | Crystal structure of uncharacterized conserved protein from Pyrococcus horikoshii | | Descriptor: | Putative uncharacterized protein PH1780, S-ADENOSYLMETHIONINE | | Authors: | Kanagawa, M, Minami, Y, Watanabe, N, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-06 | | Release date: | 2007-11-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Crystal structure of uncharacterized conserved protein from Pyrococcus horikoshii
To be Published
|
|
2ZV2
 
 | | Crystal structure of human calcium/calmodulin-dependent protein kinase kinase 2, beta, CaMKK2 kinase domain in complex with STO-609 | | Descriptor: | 7-oxo-7H-benzimidazo[2,1-a]benz[de]isoquinoline-3-carboxylic acid, Calcium/calmodulin-dependent protein kinase kinase 2 | | Authors: | Yoshikawa, S, Kukimoto-niino, M, Shirouzu, M, Suzuki, A, Lee, S, Minokoshi, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-10-31 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the Ca2+/calmodulin-dependent protein kinase kinase in complex with the inhibitor STO-609
J.Biol.Chem., 286, 2011
|
|
5ZV2
 
 | | FGFR-1 in complex with ligand lenvatinib | | Descriptor: | 4-{3-chloro-4-[(cyclopropylcarbamoyl)amino]phenoxy}-7-methoxyquinoline-6-carboxamide, Fibroblast growth factor receptor 1 | | Authors: | Matsuki, M, Hoshi, T, Yamamoto, Y, Ikemori-Kawada, M, Minoshima, Y, Funahashi, Y, Matsui, J. | | Deposit date: | 2018-05-09 | | Release date: | 2018-07-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Lenvatinib inhibits angiogenesis and tumor fibroblast growth factor signaling pathways in human hepatocellular carcinoma models.
Cancer Med, 7, 2018
|
|
5F4N
 
 | | Multi-parameter lead optimization to give an oral CHK1 inhibitor clinical candidate: (R)-5-((4-((morpholin-2-ylmethyl)amino)-5-(trifluoromethyl)pyridin-2-yl)amino)pyrazine-2-carbonitrile (CCT245737) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Serine/threonine-protein kinase Chk1, ... | | Authors: | Collins, I, Garrett, M.D, van Montfort, R, Osborne, J.D, Matthews, T.P, McHardy, T, Proisy, N, Cheung, K.J, Lainchbury, M, Brown, N, Walton, M.I, Eve, P.D, Boxall, K.J, Hayes, A, Henley, A.T, Valenti, M.R, De Haven Brandon, A.K, Box, G, Westwood, I.M, Jamin, Y, Robinson, S.P, Leonard, P, Reader, J.C, Aherne, G.W, Raynaud, F.I, Eccles, S.A. | | Deposit date: | 2015-12-03 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Multiparameter Lead Optimization to Give an Oral Checkpoint Kinase 1 (CHK1) Inhibitor Clinical Candidate: (R)-5-((4-((Morpholin-2-ylmethyl)amino)-5-(trifluoromethyl)pyridin-2-yl)amino)pyrazine-2-carbonitrile (CCT245737).
J.Med.Chem., 59, 2016
|
|
2AMN
 
 | | Solution structure of Fowlicidin-1, a novel Cathelicidin antimicrobial peptide from chicken | | Descriptor: | cathelicidin | | Authors: | Xiao, Y, Dai, H, Bommineni, Y.R, Prakash, O, Zhang, G. | | Deposit date: | 2005-08-09 | | Release date: | 2006-07-18 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Structure-activity relationships of fowlicidin-1, a cathelicidin antimicrobial peptide in chicken.
Febs J., 273, 2006
|
|
2D2A
 
 | | Crystal Structure of Escherichia coli SufA Involved in Biosynthesis of Iron-sulfur Clusters | | Descriptor: | SufA protein | | Authors: | Wada, K, Hasegawa, Y, Gong, Z, Minami, Y, Fukuyama, K, Takahashi, Y. | | Deposit date: | 2005-09-05 | | Release date: | 2005-12-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Escherichia coli SufA involved in biosynthesis of iron-sulfur clusters: Implications for a functional dimer
Febs Lett., 579, 2005
|
|
7C4N
 
 | | Ancestral L-amino acid oxidase (AncLAAO-N5) L-Phe binding form | | Descriptor: | Ancestral L-amino acid oxidase, FLAVIN-ADENINE DINUCLEOTIDE, PHENYLALANINE | | Authors: | Nakano, S, Minamino, Y, Karasuda, H, Ito, S. | | Deposit date: | 2020-05-18 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ancestral L-amino acid oxidases for deracemization and stereoinversion of amino acids
Commun Chem, 3, 2020
|
|
