6L8S
 
 | | High resolution crystal structure of crustacean hemocyanin. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Masuda, T, Mikami, B, Baba, S. | | Deposit date: | 2019-11-07 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | The high-resolution crystal structure of lobster hemocyanin shows its enzymatic capability as a phenoloxidase.
Arch.Biochem.Biophys., 688, 2020
|
|
6M1W
 
 | | Structure of the 2-Aminoisobutyric acid Monooxygenase Hydroxylase | | Descriptor: | Amidohydrolase, CHLORIDE ION, FE (III) ION, ... | | Authors: | Hibi, M, Mikami, B, Ogawa, J. | | Deposit date: | 2020-02-26 | | Release date: | 2021-01-06 | | Last modified: | 2021-01-27 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A three-component monooxygenase from Rhodococcus wratislaviensis may expand industrial applications of bacterial enzymes.
Commun Biol, 4, 2021
|
|
6M2I
 
 | | Structure of the 2-Aminoisobutyric acid Monooxygenase Hydroxylase | | Descriptor: | 1,2-ETHANEDIOL, Amidohydrolase, FE (III) ION, ... | | Authors: | Hibi, M, Mikami, B, Ogawa, J. | | Deposit date: | 2020-02-27 | | Release date: | 2021-01-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A three-component monooxygenase from Rhodococcus wratislaviensis may expand industrial applications of bacterial enzymes.
Commun Biol, 4, 2021
|
|
7YE3
 
 | | Crystal structure of Lactobacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI complexed with MES | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase, ZINC ION | | Authors: | Yamamoto, Y, Oiki, S, Takase, R, Mikami, B, Hashimoto, W. | | Deposit date: | 2022-07-05 | | Release date: | 2023-07-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | Crystal Structures of Lacticaseibacillus 4-Deoxy-L- threo- 5-hexosulose-uronate Ketol-isomerase KduI in Complex with Substrate Analogs.
J Appl Glycosci (1999), 70, 2023
|
|
7YRS
 
 | | Crystal structure of Lactobacillus rhamnosus 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase KduI complexed with MOPS | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, 4-deoxy-L-threo-5-hexosulose-uronate ketol-isomerase, ZINC ION | | Authors: | Yamamoto, Y, Oiki, S, Takase, R, Mikami, B, Hashimoto, W. | | Deposit date: | 2022-08-10 | | Release date: | 2023-08-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Crystal Structures of Lacticaseibacillus 4-Deoxy-L- threo- 5-hexosulose-uronate Ketol-isomerase KduI in Complex with Substrate Analogs.
J Appl Glycosci (1999), 70, 2023
|
|
4XIG
 
 | | Crystal structure of bacterial alginate ABC transporter determined through humid air and glue-coating method | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid, AlgM1, AlgM2, ... | | Authors: | Kaneko, A, Maruyama, Y, Mizuno, N, Baba, S, Kumasaka, T, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-01-07 | | Release date: | 2016-01-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.402 Å) | | Cite: | A solute-binding protein in the closed conformation induces ATP hydrolysis in a bacterial ATP-binding cassette transporter involved in the import of alginate.
J.Biol.Chem., 292, 2017
|
|
4XVB
 
 | |
4XTC
 
 | | Crystal structure of bacterial alginate ABC transporter in complex with alginate pentasaccharide-bound periplasmic protein | | Descriptor: | AlgM1, AlgM2, AlgQ2, ... | | Authors: | Kaneko, A, Maruyama, Y, Mizuno, N, Baba, S, Kumasaka, T, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-01-23 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | A solute-binding protein in the closed conformation induces ATP hydrolysis in a bacterial ATP-binding cassette transporter involved in the import of alginate.
J.Biol.Chem., 292, 2017
|
|
3QAC
 
 | | Structure of amaranth 11S proglobulin seed storage protein from Amaranthus hypochondriacus L. | | Descriptor: | 11S globulin seed storage protein | | Authors: | Tandang-Silvas, M.R, Carrazco-Pena, L, Barba de la Rosa, A.P, Osuna-Castro, J.A, Utsumi, S, Mikami, B, Maruyama, N. | | Deposit date: | 2011-01-10 | | Release date: | 2012-01-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.275 Å) | | Cite: | Structure of amaranth 11S proglobulin, a major seed storage protein from Amaranthus hypochondriacus L.
To be Published
|
|
3Q9T
 
 | | Crystal structure analysis of formate oxidase | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Doubayashi, D, Ootake, T, Maeda, Y, Oki, M, Tokunaga, Y, Sakurai, A, Nagaosa, Y, Mikami, B, Uchida, H. | | Deposit date: | 2011-01-09 | | Release date: | 2011-09-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Formate oxidase, an enzyme of the glucose-methanol-choline oxidoreductase family, has a His-Arg pair and 8-formyl-FAD at the catalytic site.
Biosci.Biotechnol.Biochem., 75, 2011
|
|
3SMH
 
 | |
1OVT
 
 | |
3KGL
 
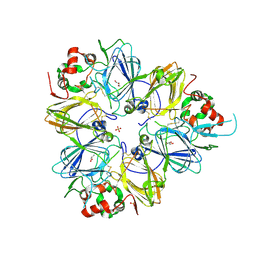 | | Crystal structure of procruciferin, 11S globulin from Brassica napus | | Descriptor: | Cruciferin, GLYCEROL, SULFATE ION | | Authors: | Tandang-Silvas, M.R, Mikami, B, Maruyama, N, Utsumi, S. | | Deposit date: | 2009-10-29 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.981 Å) | | Cite: | Conservation and divergence on plant seed 11S globulins based on crystal structures.
Biochim.Biophys.Acta, 1804, 2010
|
|
8JT1
 
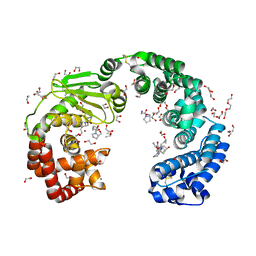 | | COLLAGENASE FROM GRIMONTIA (VIBRIO) HOLLISAE 1706B COMPLEXED WITH GLY-PRO-HYP-GLY-PRO-HYP | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 6-mer peptide, ... | | Authors: | Ueshima, S, Yaskawa, K, Takita, T, Mikami, B. | | Deposit date: | 2023-06-21 | | Release date: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into the catalytic mechanism of Grimontia hollisae collagenase through structural and mutational analyses.
Febs Lett., 597, 2023
|
|
8JZ8
 
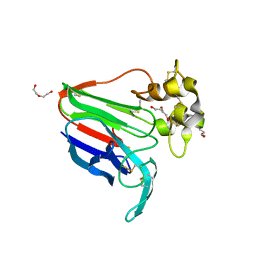 | | Subatomic structure of orthorhombic thaumatin at 0.89 Angstroms | | Descriptor: | DI(HYDROXYETHYL)ETHER, Thaumatin I | | Authors: | Masuda, T, Suzuki, M, Yamasaki, M, Mikami, B. | | Deposit date: | 2023-07-04 | | Release date: | 2024-05-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Subatomic structure of orthorhombic thaumatin at 0.89 angstrom reveals that highly flexible conformations are crucial for thaumatin sweetness.
Biochem.Biophys.Res.Commun., 703, 2024
|
|
5CL2
 
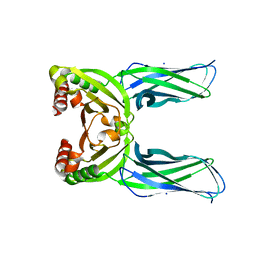 | | Crystal structure of Spo0M, sporulation control protein, from Bacillus subtilis. | | Descriptor: | SODIUM ION, Sporulation-control protein spo0M | | Authors: | Sonoda, Y, Mizutani, K, Mikami, B. | | Deposit date: | 2015-07-16 | | Release date: | 2015-12-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Spo0M, a sporulation-control protein from Bacillus subtilis.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4Z9Y
 
 | | Crystal structure of 2-keto-3-deoxy-D-gluconate dehydrogenase from Pectobacterium carotovorum | | Descriptor: | 2-deoxy-D-gluconate 3-dehydrogenase, SULFATE ION | | Authors: | Takase, R, Maruyama, Y, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-04-13 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural determinants in bacterial 2-keto-3-deoxy-D-gluconate dehydrogenase KduD for dual-coenzyme specificity
Proteins, 84, 2016
|
|
2RGK
 
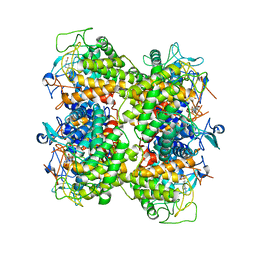 | | Functional annotation of Escherichia coli yihS-encoded protein | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Uncharacterized sugar isomerase yihS | | Authors: | Itoh, T, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-10-03 | | Release date: | 2008-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of YihS in complex with D-mannose: structural annotation of Escherichia coli and Salmonella enterica yihS-encoded proteins to an aldose-ketose isomerase
J.Mol.Biol., 377, 2008
|
|
4ZA2
 
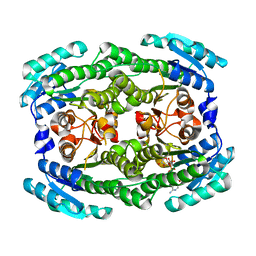 | | Crystal structure of Pectobacterium carotovorum 2-keto-3-deoxy-D-gluconate dehydrogenase complexed with NAD+ | | Descriptor: | 2-deoxy-D-gluconate 3-dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Takase, R, Maruyama, Y, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-04-13 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural determinants in bacterial 2-keto-3-deoxy-D-gluconate dehydrogenase KduD for dual-coenzyme specificity
Proteins, 84, 2016
|
|
5E4P
 
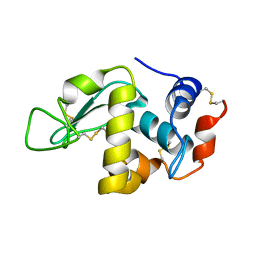 | | X-ray Crystal Structure Analysis of Magnetically Oriented Microcrystals of Lysozyme at 1.8 angstrom Resolution | | Descriptor: | Lysozyme C | | Authors: | Tsukui, S, Kimura, F, Garman, E.F, Baba, S, Mizuno, N, Mikami, B, Kimura, T. | | Deposit date: | 2015-10-06 | | Release date: | 2016-08-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | X-ray crystal structure analysis of magnetically oriented microcrystals of lysozyme at 1.8 A resolution
J.Appl.Crystallogr., 49, 2016
|
|
1FP3
 
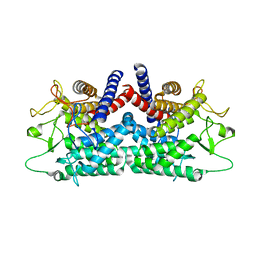 | | CRYSTAL STRUCTURE OF N-ACYL-D-GLUCOSAMINE 2-EPIMERASE FROM PORCINE KIDNEY | | Descriptor: | N-ACYL-D-GLUCOSAMINE 2-EPIMERASE | | Authors: | Itoh, T, Mikami, B, Maru, I, Ohta, Y, Hashimoto, W, Murata, K. | | Deposit date: | 2000-08-30 | | Release date: | 2000-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of N-acyl-D-glucosamine 2-epimerase from porcine kidney at 2.0 A resolution.
J.Mol.Biol., 303, 2000
|
|
4Z9X
 
 | | Crystal structure of 2-keto-3-deoxy-D-gluconate dehydrogenase from Streptococcus pyogenes | | Descriptor: | Gluconate 5-dehydrogenase | | Authors: | Maruyama, Y, Takase, R, Oiki, S, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2015-04-13 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural determinants in bacterial 2-keto-3-deoxy-D-gluconate dehydrogenase KduD for dual-coenzyme specificity
Proteins, 84, 2016
|
|
5SW0
 
 | |
5SW1
 
 | | Thaumatin Structure at pH 6.0 | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, Thaumatin I | | Authors: | Masuda, T, Sano, A, Murata, K, Okubo, K, Suzuki, M, Mikami, B. | | Deposit date: | 2016-08-08 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Thaumatin Structure at pH 6.0
To Be Published
|
|
5SW2
 
 | | Thaumatin Structure at pH 6.0, orthorhombic type1 | | Descriptor: | GLYCEROL, Thaumatin I | | Authors: | Masuda, T, Sano, A, Murata, K, Okubo, K, Suzuki, M, Mikami, B. | | Deposit date: | 2016-08-08 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Thaumatin Structure at pH 6.0, orthorhombic type1
To Be Published
|
|
