5WFI
 
 | |
1PB3
 
 | |
1PB1
 
 | |
6DHC
 
 | | X-ray structure of BACE1 in complex with a bicyclic isoxazoline carboxamide as the P3 ligand | | 分子名称: | (3R,3aR,6aS)-N-[(4R,7S,8S,10R,13S)-8-hydroxy-10,17-dimethyl-7-(2-methylpropyl)-5,11,14-trioxo-13-(propan-2-yl)-2-thia-6,12,15-triazaoctadecan-4-yl]hexahydrofuro[3,2-d][1,2]oxazole-3-carboxamide, Beta-secretase 1, GLYCEROL, ... | | 著者 | Mesecar, A.D, Lendy, E.K. | | 登録日 | 2018-05-19 | | 公開日 | 2018-07-25 | | 最終更新日 | 2020-01-01 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Design, synthesis, X-ray studies, and biological evaluation of novel BACE1 inhibitors with bicyclic isoxazoline carboxamides as the P3 ligand.
Bioorg. Med. Chem. Lett., 28, 2018
|
|
7RC0
 
 | | X-ray Structure of SARS-CoV-2 main protease covalently modified by compound GRL-091-20 | | 分子名称: | 3C-like proteinase, 5-chloro-4-methylpyridin-3-yl 1H-indole-4-carboxylate, SODIUM ION | | 著者 | Mesecar, A.D, Anson, B.A, Ghosh, A.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-07-06 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Indole Chloropyridinyl Ester-Derived SARS-CoV-2 3CLpro Inhibitors: Enzyme Inhibition, Antiviral Efficacy, Structure-Activity Relationship, and X-ray Structural Studies.
J.Med.Chem., 64, 2021
|
|
7RC1
 
 | | X-ray Structure of SARS-CoV main protease covalently modified by compound GRL-0686 | | 分子名称: | 3C-like proteinase, 5-chloropyridin-3-yl 1-(3-nitrobenzene-1-sulfonyl)-1H-indole-5-carboxylate, DIMETHYL SULFOXIDE | | 著者 | Mesecar, A.D, Anson, B.A, Ghosh, A.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-07-07 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | Indole Chloropyridinyl Ester-Derived SARS-CoV-2 3CLpro Inhibitors: Enzyme Inhibition, Antiviral Efficacy, Structure-Activity Relationship, and X-ray Structural Studies.
J.Med.Chem., 64, 2021
|
|
7RBZ
 
 | | X-ray Structure of SARS-CoV-2 main protease covalently modified by compound GRL-017-20 | | 分子名称: | 3C-like proteinase, 5-chloropyridin-3-yl 2,3-dihydro-1H-indole-4-carboxylate | | 著者 | Mesecar, A.D, Anson, B.A, Ghosh, A.K, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-07-06 | | 公開日 | 2021-09-29 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Indole Chloropyridinyl Ester-Derived SARS-CoV-2 3CLpro Inhibitors: Enzyme Inhibition, Antiviral Efficacy, Structure-Activity Relationship, and X-ray Structural Studies.
J.Med.Chem., 64, 2021
|
|
1P8F
 
 | |
1QW7
 
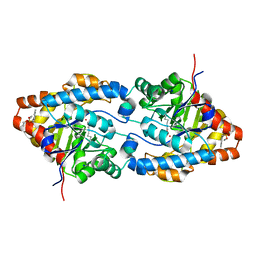 | | Structure of an Engineered Organophosphorous Hydrolase with Increased Activity Toward Hydrolysis of Phosphothiolate Bonds | | 分子名称: | COBALT (II) ION, DIETHYL 4-METHYLBENZYLPHOSPHONATE, Parathion hydrolase, ... | | 著者 | Mesecar, A.D, Grimsley, J.K, Holton, T, Wild, J.R. | | 登録日 | 2003-09-01 | | 公開日 | 2004-11-30 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and mutational studies of organophosphorus hydrolase reveal a cryptic and functional allosteric-binding site.
Arch.Biochem.Biophys., 442, 2005
|
|
1TX8
 
 | | Bovine Trypsin complexed with AMSO | | 分子名称: | 4-(METHYLSULFONYL)BENZENECARBOXIMIDAMIDE, CALCIUM ION, Trypsinogen | | 著者 | Mesecar, A.D. | | 登録日 | 2004-07-02 | | 公開日 | 2005-10-18 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Design, synthesis, and evaluation of oxyanion-hole selective inhibitor substituents for the S1 subsite of factor Xa
Bioorg.Med.Chem.Lett., 14, 2004
|
|
4M5X
 
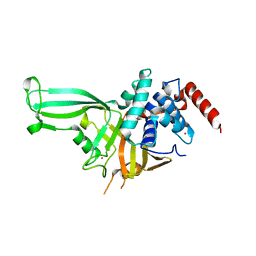 | | Crystal structure of the USP7/HAUSP catalytic domain | | 分子名称: | BROMIDE ION, Ubiquitin carboxyl-terminal hydrolase 7 | | 著者 | Mesecar, A.D, Molland, K.L, Zhou, Q. | | 登録日 | 2013-08-08 | | 公開日 | 2014-03-12 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.187 Å) | | 主引用文献 | A 2.2 angstrom resolution structure of the USP7 catalytic domain in a new space group elaborates upon structural rearrangements resulting from ubiquitin binding.
Acta Crystallogr F Struct Biol Commun, 70, 2014
|
|
4MDS
 
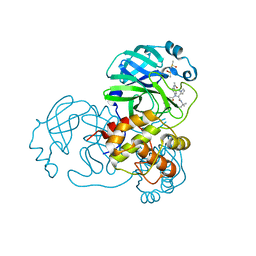 | | Discovery of N-(benzo[1,2,3]triazol-1-yl)-N-(benzyl)acetamido)phenyl) carboxamides as severe acute respiratory syndrome coronavirus (SARS-CoV) 3CLpro inhibitors: identification of ML300 and non-covalent nanomolar inhibitors with an induced-fit binding | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[4-(acetylamino)phenyl]-2-(1H-benzotriazol-1-yl)-N-[(1R)-2-[(2-methylbutan-2-yl)amino]-1-(1-methyl-1H-pyrrol-2-yl)-2-oxoethyl]acetamide | | 著者 | Mesecar, A.D, Grum-Tokars, V. | | 登録日 | 2013-08-23 | | 公開日 | 2013-10-02 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (1.598 Å) | | 主引用文献 | Discovery of N-(benzo[1,2,3]triazol-1-yl)-N-(benzyl)acetamido)phenyl) carboxamides as severe acute respiratory syndrome coronavirus (SARS-CoV) 3CLpro inhibitors: Identification of ML300 and noncovalent nanomolar inhibitors with an induced-fit binding.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
6W63
 
 | |
6W81
 
 | |
6WCO
 
 | |
6W79
 
 | |
3MJ5
 
 | | Severe Acute Respiratory Syndrome-Coronavirus Papain-Like Protease Inhibitors: Design, Synthesis, Protein-Ligand X-ray Structure and Biological Evaluation | | 分子名称: | N-(1,3-benzodioxol-5-ylmethyl)-1-[(1R)-1-naphthalen-1-ylethyl]piperidine-4-carboxamide, Replicase polyprotein 1a, ZINC ION | | 著者 | Mesecar, A.D, Ratia, K.M, Pegan, S.D. | | 登録日 | 2010-04-12 | | 公開日 | 2010-06-30 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.63 Å) | | 主引用文献 | Severe acute respiratory syndrome coronavirus papain-like novel protease inhibitors: design, synthesis, protein-ligand X-ray structure and biological evaluation
J.Med.Chem., 53, 2010
|
|
3E9S
 
 | |
4MM3
 
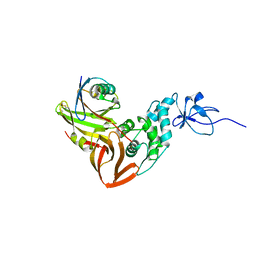 | |
3MB2
 
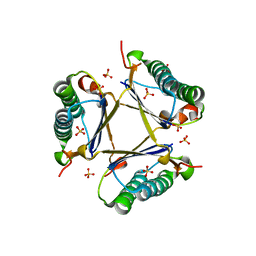 | | Kinetic and Structural Characterization of a Heterohexamer 4-Oxalocrotonate Tautomerase from Chloroflexus aurantiacus J-10-fl: Implications for Functional and Structural Diversity in the Tautomerase Superfamily | | 分子名称: | 4-oxalocrotonate tautomerase family enzyme - alpha subunit, 4-oxalocrotonate tautomerase family enzyme - beta subunit, SULFATE ION | | 著者 | Burks, E.A, Fleming, C.D, Mesecar, A.D, Whitman, C.P, Pegan, S.D. | | 登録日 | 2010-03-24 | | 公開日 | 2010-06-02 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (2.41 Å) | | 主引用文献 | Kinetic and structural characterization of a heterohexamer 4-oxalocrotonate tautomerase from Chloroflexus aurantiacus J-10-fl: implications for functional and structural diversity in the tautomerase superfamily
Biochemistry, 49, 2010
|
|
5HYO
 
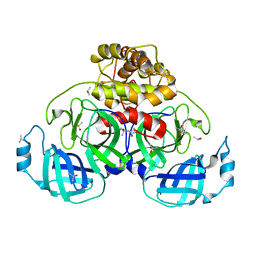 | |
4ZRO
 
 | | 2.1 A X-Ray Structure of FIPV-3CLpro bound to covalent inhibitor | | 分子名称: | 3C-like proteinase, Bounded inhibitor of N-(tert-butoxycarbonyl)-L-seryl-L-valyl-N-{(2S)-5-ethoxy-5-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]pentan-2-yl}-L-leucinamide, DIMETHYL SULFOXIDE | | 著者 | St John, S.E, Mesecar, A.D. | | 登録日 | 2015-05-12 | | 公開日 | 2015-10-14 | | 最終更新日 | 2019-12-11 | | 実験手法 | X-RAY DIFFRACTION (2.0566 Å) | | 主引用文献 | X-ray structure and inhibition of the feline infectious peritonitis virus 3C-like protease: Structural implications for drug design.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
2ZC1
 
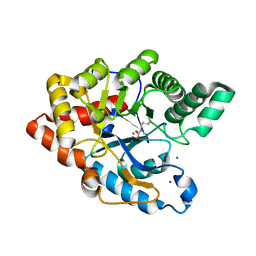 | | Organophosphorus Hydrolase from Deinococcus radiodurans | | 分子名称: | BROMIDE ION, COBALT (II) ION, Phosphotriesterase | | 著者 | Larsen, S.D, Hawwa, R, Ratia, K, Santarsiero, B.D, Mesecar, A.D. | | 登録日 | 2007-11-02 | | 公開日 | 2008-11-25 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | X-Ray Structural Insights into a Phosphotriesterase
to be published
|
|
5KO3
 
 | |
6UJ0
 
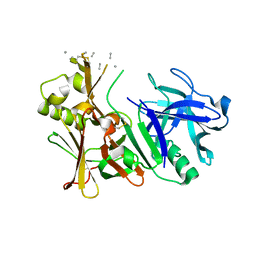 | | Unbound BACE2 mutant structure | | 分子名称: | Beta-secretase 2, unidentified polypeptide | | 著者 | Yen, Y.C, Ghosh, A.K, Mesecar, A.D. | | 登録日 | 2019-10-01 | | 公開日 | 2020-10-07 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | A Structure-Based Discovery Platform for BACE2 and the Development of Selective BACE Inhibitors.
Acs Chem Neurosci, 12, 2021
|
|
