8UPF
 
 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168-UbcH5c | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-10-22 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8TXV
 
 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A K15 in complex with RNF168 (Class 1) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8TXW
 
 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A K15 in complex with RNF168 (Class 2) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
6CO2
 
 | | Structure of an engineered protein (NUDT16TI) in complex with 53BP1 Tudor domains | | Descriptor: | NUDT16-Tudor-interacting (NUDT16TI), TP53-binding protein 1 | | Authors: | Botuyan, M.V, Thompson, J.R, Cui, G, Mer, G. | | Deposit date: | 2018-03-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Mechanism of 53BP1 activity regulation by RNA-binding TIRR and a designer protein.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6CO1
 
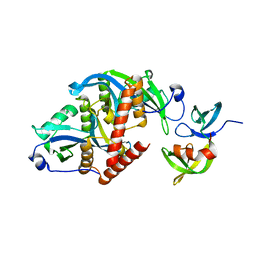 | |
6D0L
 
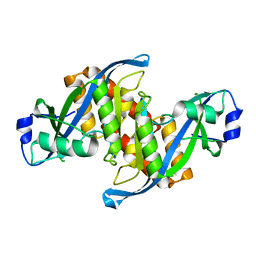 | | Structure of human TIRR | | Descriptor: | Tudor-interacting repair regulator protein | | Authors: | Cui, G, Botuyan, M.V, Mer, G. | | Deposit date: | 2018-04-10 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mechanism of 53BP1 activity regulation by RNA-binding TIRR and a designer protein.
Nat. Struct. Mol. Biol., 25, 2018
|
|
7LYC
 
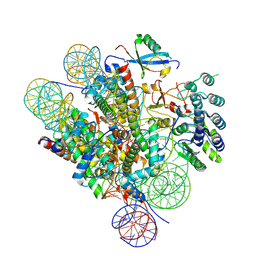 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A Lys13 and Lys15 in complex with BARD1 (residues 415-777) | | Descriptor: | BRCA1-associated RING domain protein 1, DNA (146-MER), DNA (147-MER), ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, D, Mer, E, Mer, G. | | Deposit date: | 2021-03-06 | | Release date: | 2021-06-16 | | Last modified: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Mechanisms of BRCA1-BARD1 nucleosome recognition and ubiquitylation.
Nature, 596, 2021
|
|
7LYA
 
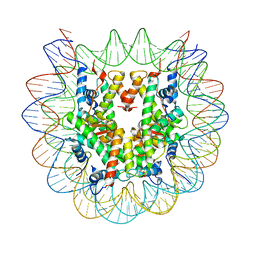 | | Cryo-EM structure of the human nucleosome core particle with linked histone proteins H2A and H2B | | Descriptor: | DNA (146-MER), DNA (147-MER), Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, D, Mer, E, Mer, G. | | Deposit date: | 2021-03-06 | | Release date: | 2021-07-28 | | Last modified: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Mechanisms of BRCA1-BARD1 nucleosome recognition and ubiquitylation.
Nature, 596, 2021
|
|
7LYB
 
 | | Cryo-EM structure of the human nucleosome core particle in complex with BRCA1-BARD1-UbcH5c | | Descriptor: | BRCA1-associated RING domain protein 1, DNA (146-MER), DNA (147-MER), ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, D, Mer, E, Mer, G. | | Deposit date: | 2021-03-06 | | Release date: | 2021-07-28 | | Last modified: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Mechanisms of BRCA1-BARD1 nucleosome recognition and ubiquitylation.
Nature, 596, 2021
|
|
8U4U
 
 | |
8UQD
 
 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 20-residue linker (condition 2. RING not modeled in density) | | Descriptor: | E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.893 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQC
 
 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 20-residue linker (crystallization condition 2) | | Descriptor: | E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, ZINC ION | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQB
 
 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 20-residue linker (crystallization condition 1) | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, ZINC ION | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.484 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQE
 
 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 26-residue linker (RING not modeled in density) | | Descriptor: | E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.562 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQA
 
 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 12-residue linker | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, SODIUM ION, ... | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQ9
 
 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 4-residue linker | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, GLYCEROL, ... | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQ8
 
 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 2-residue linker | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, GLYCEROL, ... | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
5VZM
 
 | |
6VE5
 
 | |
3TW1
 
 | | Structure of Rtt106-AHN | | Descriptor: | GLYCEROL, Histone chaperone RTT106, N-[2-(1H-IMIDAZOL-4-YL)ETHYL]ACETAMIDE | | Authors: | Su, D, Thompson, J.R, Mer, G. | | Deposit date: | 2011-09-21 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.772 Å) | | Cite: | Structural basis for recognition of H3K56-acetylated histone H3-H4 by the chaperone Rtt106.
Nature, 483, 2012
|
|
3TVV
 
 | |
5VF0
 
 | |
3P8D
 
 | | Crystal structure of the second Tudor domain of human PHF20 (homodimer form) | | Descriptor: | Medulloblastoma antigen MU-MB-50.72 | | Authors: | Cui, G, Lee, J, Thompson, J.R, Botuyan, M.V, Mer, G. | | Deposit date: | 2010-10-13 | | Release date: | 2011-06-22 | | Last modified: | 2012-09-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | PHF20 is an effector protein of p53 double lysine methylation that stabilizes and activates p53.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3PD7
 
 | |
3SD4
 
 | |
