8AXC
 
 | | Crystal structure of mouse Ces2c | | Descriptor: | Acylcarnitine hydrolase, CHLORIDE ION, NICOTINAMIDE, ... | | Authors: | Eisner, H, Riegler-Berket, L, Rodriguez Gamez, C, Sagmeister, T, Chalhoub, G, Darnhofer, B, Panikkaveetil Jawaharlal, J, Birner-Gruenberger, R, Pavkov-Keller, T, Haemmerle, G, Schoiswohl, G, Oberer, M. | | Deposit date: | 2022-08-31 | | Release date: | 2022-11-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | The Crystal Structure of Mouse Ces2c, a Potential Ortholog of Human CES2, Shows Structural Similarities in Substrate Regulation and Product Release to Human CES1.
Int J Mol Sci, 23, 2022
|
|
8OID
 
 | | Cryo-EM structure of ADP-bound, filamentous beta-actin harboring the N111S mutation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Oosterheert, W, Blanc, F.E.C, Roy, A, Belyy, A, Hofnagel, O, Hummer, G, Bieling, P, Raunser, S. | | Deposit date: | 2023-03-22 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-22 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Molecular mechanisms of inorganic-phosphate release from the core and barbed end of actin filaments.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4KI8
 
 | | Crystal structure of a GroEL-ADP complex in the relaxed allosteric state | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, ... | | Authors: | Fei, X, Yang, D, LaRonde-LeBlanc, N, Lorimer, G.H. | | Deposit date: | 2013-05-01 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.722 Å) | | Cite: | Crystal structure of a GroEL-ADP complex in the relaxed allosteric state at 2.7 A resolution.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4OH2
 
 | | Crystal Structure of Cu/Zn Superoxide Dismutase I149T | | Descriptor: | COPPER (II) ION, Superoxide dismutase [Cu-Zn], ZINC ION | | Authors: | Crane, B.R, Merz, G.E. | | Deposit date: | 2014-01-16 | | Release date: | 2014-10-15 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.384 Å) | | Cite: | Copper-Based Pulsed Dipolar ESR Spectroscopy as a Probe of Protein Conformation Linked to Disease States.
Biophys.J., 107, 2014
|
|
4PKN
 
 | | Crystal structure of the football-shaped GroEL-GroES2-(ADPBeFx)14 complex containing substrate Rubisco | | Descriptor: | 10 kDa chaperonin, 60 kDa chaperonin, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Fei, X, Ye, X, Laronde-Leblanc, N, Lorimer, G.H. | | Deposit date: | 2014-05-15 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.66 Å) | | Cite: | Formation and structures of GroEL:GroES2 chaperonin footballs, the protein-folding functional form.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
6CGV
 
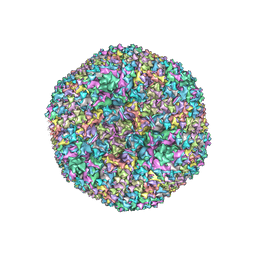 | | Revised crystal structure of human adenovirus | | Descriptor: | Hexon protein, Hexon-interlacing protein, Penton protein, ... | | Authors: | Natchiar, S.K, Venkataraman, S, Nemerow, G.R, Reddy, V.S. | | Deposit date: | 2018-02-21 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Revised Crystal Structure of Human Adenovirus Reveals the Limits on Protein IX Quasi-Equivalence and on Analyzing Large Macromolecular Complexes.
J. Mol. Biol., 430, 2018
|
|
2Z9T
 
 | | Crystal structure of the human beta-2 microglobulin mutant W60G | | Descriptor: | Beta-2-microglobulin | | Authors: | Ricagno, S, Bolognesi, M, Bellotti, V, Corazza, A, Rennella, E, Gural, D, Mimmi, M.C, Betto, E, Pucillo, C, Fogolari, F, Viglino, P, Raimondi, S, Giorgetti, S, Bolognesi, B, Merlini, G, Stoppini, M. | | Deposit date: | 2007-09-26 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The controlling roles of Trp60 and Trp95 in beta2-microglobulin function, folding and amyloid aggregation properties
J.Mol.Biol., 378, 2008
|
|
6EJJ
 
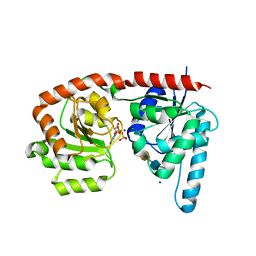 | | Structure of a glycosyltransferase / state 2 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-glucopyranose, CHLORIDE ION, NerylNeryl pyrophosphate, ... | | Authors: | Ramirez, A.S, Boilevin, J, Mehdipour, A.R, Hummer, G, Darbre, T, Reymond, J.L, Locher, K.P. | | Deposit date: | 2017-09-21 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of the molecular ruler mechanism of a bacterial glycosyltransferase.
Nat Commun, 9, 2018
|
|
3E76
 
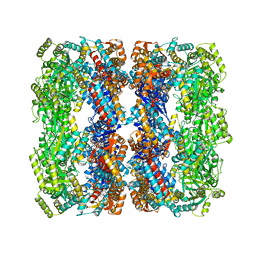 | | Crystal structure of Wild-type GroEL with bound Thallium ions | | Descriptor: | 60 kDa chaperonin, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Kiser, P.D, Lorimer, G.H, Palczewski, K. | | Deposit date: | 2008-08-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.94 Å) | | Cite: | Use of thallium to identify monovalent cation binding sites in GroEL.
Acta Crystallogr.,Sect.F, 65, 2009
|
|
2BM2
 
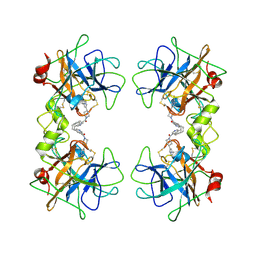 | | human beta-II tryptase in complex with 4-(3-Aminomethyl-phenyl)- piperidin-1-yl-(5-phenethyl- pyridin-3-yl)-methanone | | Descriptor: | 1-[3-(1-{[5-(2-PHENYLETHYL)PYRIDIN-3-YL]CARBONYL}PIPERIDIN-4-YL)PHENYL]METHANAMINE, HUMAN BETA2 TRYPTASE | | Authors: | Maignan, S, Guilloteau, J.-P, Dupuy, A, Levell, J, Astles, P, Eastwood, P, Cairns, J, Houille, O, Aldous, S, Merriman, G, Whiteley, B, Pribish, J, Czekaj, M, Liang, G, Davidson, J, Harrison, T, Morley, A, Watson, S, Fenton, G, Mccarthy, C, Romano, J, Mathew, R, Engers, D, Gardyan, M, Sides, K, Kwong, J, Tsay, J, Rebello, S, Shen, L, Wang, J, Luo, Y, Giardino, O, Lim, H.-K, Smith, K, Pauls, H. | | Deposit date: | 2005-03-09 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure Based Design of 4-(3-Aminomethylphenyl) Piperidinyl-1-Amides: Novel, Potent, Selective, and Orally Bioavailable Inhibitors of Bii Tryptase
Bioorg.Med.Chem., 13, 2005
|
|
3CNC
 
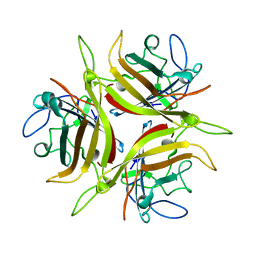 | |
4AFJ
 
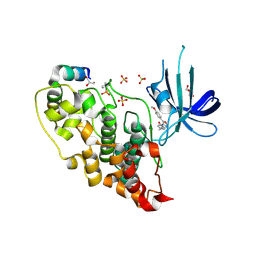 | | 5-aryl-4-carboxamide-1,3-oxazoles: potent and selective GSK-3 inhibitors | | Descriptor: | 5-(4-METHOXYPHENYL)-N-(PYRIDIN-4-YLMETHYL)-1,3-OXAZOLE-4-CARBOXAMIDE, GLYCEROL, GLYCOGEN SYNTHASE KINASE-3 BETA, ... | | Authors: | Gentile, G, Merlo, G, Pozzan, A, Bernasconi, G, Bax, B, Bamborough, P, Bridges, A, Carter, P, Neu, M, Yao, G, Brough, C, Cutler, G, Coffin, A, Belyanskaya, S. | | Deposit date: | 2012-01-19 | | Release date: | 2012-02-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | 5-Aryl-4-Carboxamide-1,3-Oxazoles: Potent and Selective Gsk-3 Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
2VU2
 
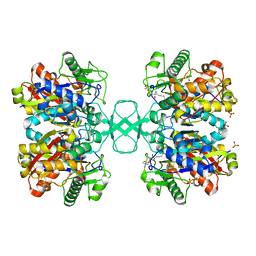 | | Biosynthetic thiolase from Z. ramigera. Complex with S-pantetheine-11- pivalate. | | Descriptor: | (3R)-3-hydroxy-2,2-dimethyl-4-oxo-4-({3-oxo-3-[(2-sulfanylethyl)amino]propyl}amino)butyl 2,2-dimethylpropanoate, ACETYL-COA ACETYLTRANSFERASE, SULFATE ION | | Authors: | Kursula, P, Merilainen, G, Schmitz, W, Wierenga, R.K. | | Deposit date: | 2008-05-19 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Sulfur Atoms of the Substrate Coa and the Catalytic Cysteine are Required for a Productive Mode of Substrate Binding in Bacterial Biosynthetic Thiolase, a Thioester-Dependent Enzyme.
FEBS J., 275, 2008
|
|
2VB5
 
 | | Solution structure of W60G mutant of human beta2-microglobulin | | Descriptor: | BETA-2-MICROGLOBULIN | | Authors: | Esposito, G, Corazza, A, Rennella, E, Gumral, D, Mimmi, M.C, Fogolari, F, Viglino, P, Raimondi, S, Giorgetti, S, Bolognesi, B, Merlini, G, Stoppini, M, Bellotti, V. | | Deposit date: | 2007-09-06 | | Release date: | 2007-09-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | The Controlling Roles of Trp60 and Trp95 in Beta2-Microglobulin Function, Folding and Amyloid Aggregation Properties.
J.Mol.Biol., 378, 2008
|
|
2VTZ
 
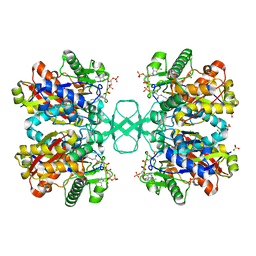 | | Biosynthetic thiolase from Z. ramigera. Complex of the C89A mutant with coenzyme A. | | Descriptor: | ACETYL-COA ACETYLTRANSFERASE, COENZYME A, SULFATE ION | | Authors: | Kursula, P, Merilainen, G, Wierenga, R.K. | | Deposit date: | 2008-05-19 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Sulfur Atoms of the Substrate Coa and the Catalytic Cysteine are Required for a Productive Mode of Substrate Binding in Bacterial Biosynthetic Thiolase, a Thioester-Dependent Enzyme.
FEBS J., 275, 2008
|
|
7KKJ
 
 | | Structure of anti-SARS-CoV-2 Spike nanobody mNb6 | | Descriptor: | CHLORIDE ION, SULFATE ION, Synthetic nanobody mNb6 | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
4BBV
 
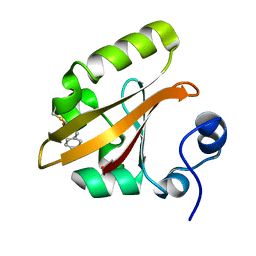 | | The PB0 Photocycle Intermediate of Photoactive Yellow Protein | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Schotte, F, Cho, H.S, Kaila, V.R.I, Kamikubo, H, Dashdorj, N, Henry, E.R, Graber, T.J, Henning, R, Wulff, M, Hummer, G, Kataoka, M, Anfinrud, P.A. | | Deposit date: | 2012-09-28 | | Release date: | 2012-11-14 | | Last modified: | 2019-01-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Watching a Signaling Protein Function in Real Time Via 100-Ps Time-Resolved Laue Crystallography.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4BBU
 
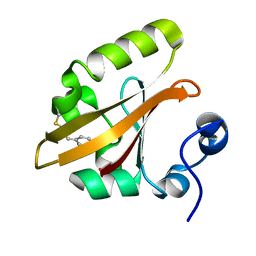 | | The PR2 Photocycle Intermediate of Photoactive Yellow Protein | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Schotte, F, Cho, H.S, Kaila, V.R.I, Kamikubo, H, Dashdorj, N, Henry, E.R, Graber, T.J, Henning, R, Wulff, M, Hummer, G, Kataoka, M, Anfinrud, P.A. | | Deposit date: | 2012-09-27 | | Release date: | 2012-11-14 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Watching a Signaling Protein Function in Real Time Via 100-Ps Time-Resolved Laue Crystallography.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4B9O
 
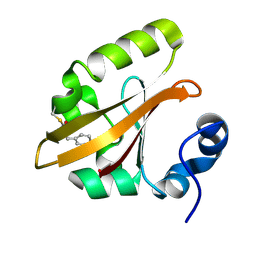 | | The PR0 Photocycle Intermediate of Photoactive Yellow Protein | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Schotte, F, Cho, H.S, Kaila, V.R.I, Kamikubo, H, Dashdorj, N, Henry, E.R, Graber, T.J, Henning, R, Wulff, M, Hummer, G, Kataoka, M, Anfinrud, P.A. | | Deposit date: | 2012-09-06 | | Release date: | 2012-11-14 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Watching a Signaling Protein Function in Real Time Via 100-Ps Time-Resolved Laue Crystallography
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4BBT
 
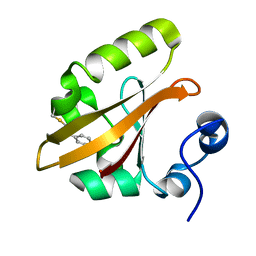 | | The PR1 Photocycle Intermediate of Photoactive Yellow Protein | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | Authors: | Schotte, F, Cho, H.S, Kaila, V.R.I, Kamikubo, H, Dashdorj, N, Henry, E.R, Graber, T.J, Henning, R, Wulff, M, Hummer, G, Kataoka, M, Anfinrud, P.A. | | Deposit date: | 2012-09-27 | | Release date: | 2012-11-14 | | Last modified: | 2019-01-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Watching a Signaling Protein Function in Real Time Via 100-Ps Time-Resolved Laue Crystallography.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3F1I
 
 | | Human ESCRT-0 Core Complex | | Descriptor: | Hepatocyte growth factor-regulated tyrosine kinase substrate, Signal transducing adapter molecule 1 | | Authors: | Ren, X, Kloer, D.P, Kim, Y, Ghirlando, R, Saidi, L, Hummer, G, Hurley, J.H. | | Deposit date: | 2008-10-28 | | Release date: | 2009-03-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Hybrid Structural Model of the Complete Human ESCRT-0 Complex.
Structure, 17, 2009
|
|
7KKK
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody Nb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KKL
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody mNb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
3PFQ
 
 | | Crystal Structure and Allosteric Activation of Protein Kinase C beta II | | Descriptor: | CALCIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Protein kinase C beta type, ... | | Authors: | Leonard, T.A, Rozycki, B, Saidi, L.F, Hummer, G, Hurley, J.H. | | Deposit date: | 2010-10-28 | | Release date: | 2011-02-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal Structure and Allosteric Activation of Protein Kinase C beta II
Cell(Cambridge,Mass.), 144, 2011
|
|
2I46
 
 | | Crystal structure of human TPP1 | | Descriptor: | Adrenocortical dysplasia protein homolog | | Authors: | Wang, F, Podell, E.R, Zaug, A.J, Yang, Y.T, Baciu, P, Else, T, Hammer, G.D, Cech, T.R, Lei, M. | | Deposit date: | 2006-08-21 | | Release date: | 2006-11-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The POT1-TPP1 telomere complex is a telomerase processivity factor.
Nature, 445, 2007
|
|
