8E1W
 
 | | Neutron crystal structure of Panus similis AA9A at room temperature | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Meilleur, F, Tandrup, T, Lo Leggio, L. | | Deposit date: | 2022-08-11 | | Release date: | 2023-01-11 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (2.1 Å), X-RAY DIFFRACTION | | Cite: | Joint X-ray/neutron structure of Lentinus similis AA9_A at room temperature.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
8DYK
 
 | |
9CXR
 
 | | X-ray Crystallographic Structure of the Poly(Hexamethylene Adipamide) (Nylon66) Hydrolase Nyl50 at Room Temperature | | Descriptor: | 1,2-ETHANEDIOL, Poly(hexamethylene adipamide) hydrolase, SODIUM ION, ... | | Authors: | Meilleur, F, Williams, A.N, Drufva, E.E, Parks, J.M, Chen, S.H, Michener, J.K. | | Deposit date: | 2024-07-31 | | Release date: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification and characterization of substrate- and product-selective nylon hydrolases.
Biorxiv, 2024
|
|
8RBN
 
 | |
8RBR
 
 | |
1YRD
 
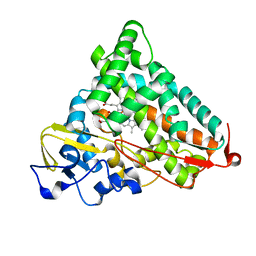 | | X-ray crystal structure of PERDEUTERATED Cytochrome P450cam | | Descriptor: | CAMPHOR, Cytochrome P450-cam, POTASSIUM ION, ... | | Authors: | Meilleur, F, Dauvergne, M.-T, Schlichting, I, Myles, D.A.A. | | Deposit date: | 2005-02-03 | | Release date: | 2005-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Production and X-ray crystallographic analysis of fully deuterated cytochrome P450cam.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1YRC
 
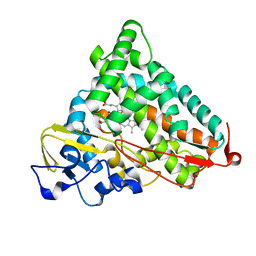 | | X-ray Crystal Structure of hydrogenated Cytochrome P450cam | | Descriptor: | CAMPHOR, Cytochrome P450-cam, POTASSIUM ION, ... | | Authors: | Meilleur, F, Dauvergne, M.-T, Schlichting, I, Myles, D.A.A. | | Deposit date: | 2005-02-03 | | Release date: | 2005-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Production and X-ray crystallographic analysis of fully deuterated cytochrome P450cam.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2PLL
 
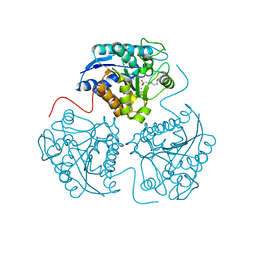 | | Crystal structure of perdeuterated human arginase I | | Descriptor: | 2(S)-AMINO-6-BORONOHEXANOIC ACID, MANGANESE (II) ION, arginase-1 | | Authors: | Di Costanzo, L, Moulin, M, Haertlein, M, Meilleur, F, Christianson, D.W. | | Deposit date: | 2007-04-19 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Expression, purification, assay, and crystal structure of perdeuterated human arginase I
Arch.Biochem.Biophys., 465, 2007
|
|
7L74
 
 | | Crystal structure of Beta-hexosyl transferase from Hamamotoa (Sporobolomyces) singularis bound to TRIS | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-hexosyltransferase, ... | | Authors: | Dagher, S.F, Edwards, B.F.P, Meilleur, F, Bruno-Barcena, J.M. | | Deposit date: | 2020-12-25 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure and mutagenic analysis of the Beta-hexosyltransferase from Hamamotoa (Sporobolomyces) singularis
To Be Published
|
|
5VNQ
 
 | | Neutron crystallographic structure of perdeuterated T4 lysozyme cysteine-free pseudo-wild type at cryogenic temperature | | Descriptor: | CHLORIDE ION, Endolysin | | Authors: | Li, L, Shukla, S, Meilleur, F, Standaert, R.F, Pierce, J, Myles, D.A.A, Cuneo, M.J. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | NEUTRON DIFFRACTION (2.2 Å) | | Cite: | Neutron crystallographic studies of T4 lysozyme at cryogenic temperature.
Protein Sci., 26, 2017
|
|
5VNR
 
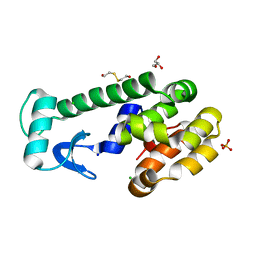 | | X-ray structure of perdeuterated T4 lysozyme cysteine-free pseudo-wild type at cryogenic temperature | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, Endolysin, ... | | Authors: | Li, L, Shukla, S, Meilleur, F, Standaert, R.F, Pierce, J, Myles, D.A.A, Cuneo, M.J. | | Deposit date: | 2017-05-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Neutron crystallographic studies of T4 lysozyme at cryogenic temperature.
Protein Sci., 26, 2017
|
|
8TT9
 
 | | X-ray structure of Macrophage Migration Inhibitory Factor (MIF) Covalently Bound to 4-hydroxyphenylpyruvate (HPP) | | Descriptor: | 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, ISOPROPYL ALCOHOL, Macrophage migration inhibitory factor | | Authors: | Schroder, G.C, Meilleur, F, Nix, J.C, Crichlow, G.V, Lolis, E.J. | | Deposit date: | 2023-08-13 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | X-ray structure of Macrophage Migration Inhibitory Factor (MIF) Covalently Bound to 4-hydroxyphenylpyruvate (HPP)
To Be Published
|
|
4RSG
 
 | | Neutron crystal structure of Ras bound to the GTP analogue GppNHp | | Descriptor: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Knihtila, R.R, Holzapfel, G, Weiss, K.L, Meilleur, F, Mattos, C. | | Deposit date: | 2014-11-07 | | Release date: | 2015-11-04 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (1.907 Å) | | Cite: | Neutron Crystal Structure of RAS GTPase Puts in Question the Protonation State of the GTP gamma-Phosphate.
J.Biol.Chem., 290, 2015
|
|
5KWF
 
 | | Joint X-ray Neutron Structure of Cholesterol Oxidase | | Descriptor: | Cholesterol oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Golden, E, Vrielink, A, Meilleur, F, Blakeley, M. | | Deposit date: | 2016-07-18 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-06 | | Method: | NEUTRON DIFFRACTION (1.499 Å), X-RAY DIFFRACTION | | Cite: | An extended N-H bond, driven by a conserved second-order interaction, orients the flavin N5 orbital in cholesterol oxidase.
Sci Rep, 7, 2017
|
|
2QXW
 
 | | Perdeuterated alr2 in complex with idd594 | | Descriptor: | Aldose reductase, CITRIC ACID, IDD594, ... | | Authors: | Blakeley, M.P, Ruiz, F, Cachau, R, Hazemann, I, Meilleur, F, Mitschler, A, Ginell, S, Afonine, P, Ventura, O, Cousido-Siah, A, Joachimiak, A, Myles, D, Podjarny, A. | | Deposit date: | 2007-08-13 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Quantum model of catalysis based on a mobile proton revealed by subatomic x-ray and neutron diffraction studies of h-aldose reductase.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2R24
 
 | | Human Aldose Reductase structure | | Descriptor: | Aldose reductase, IDD594, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Blakeley, M.P, Ruiz, F, Cachau, R, Hazemann, I, Meilleur, F, Mitschler, A, Ginell, S, Afonine, P, Ventura, O.N, Cousido-Siah, A, Haertlein, M, Joachimiak, A, Myles, D, Podjarny, A. | | Deposit date: | 2007-08-24 | | Release date: | 2008-12-23 | | Last modified: | 2024-02-21 | | Method: | NEUTRON DIFFRACTION (1.752 Å), X-RAY DIFFRACTION | | Cite: | Quantum model of catalysis based on mobile proton revealed by subatomic X-Ray and neutron diffraction studies of h-Aldose Reductase
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3SS2
 
 | | Neutron structure of perdeuterated rubredoxin using 48 hours 3rd pass data | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Munshi, P, Chung, C.-L, Blakeley, M.P, Weiss, K.L, Myles, D.A.A, Meilleur, F. | | Deposit date: | 2011-07-07 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | NEUTRON DIFFRACTION (1.75 Å) | | Cite: | Rapid visualization of hydrogen positions in protein neutron crystallographic structures.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
9DYS
 
 | |
4P1J
 
 | | Crystal structure of wild type Hypocrea jecorina Cel7a in a hexagonal crystal form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Exoglucanase 1, SAMARIUM (III) ION, ... | | Authors: | Bodenheimer, A.B, Cuneo, M.J, Swartz, P.D, Myles, D.A, Meilleur, F. | | Deposit date: | 2014-02-26 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Crystal structure of wild type Hypocrea jecorina Cel7a in a hexagonal crystal form
To Be Published
|
|
4P1H
 
 | | Crystal structure of wild type Hypocrea jecorina Cel7a in a monoclinic crystal form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BENZAMIDINE, Exoglucanase 1, ... | | Authors: | Bodenheimer, A.B, Cuneo, M.J, Swartz, P.D, Myles, D.A, Meilleur, F. | | Deposit date: | 2014-02-26 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of wild type Hypocrea jecorina Cel7a in a monoclinic crystal form
to be published
|
|
4LNC
 
 | | Neutron structure of the cyclic glucose bound Xylose Isomerase E186Q mutant | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, Xylose isomerase, ... | | Authors: | Munshi, P, Meilleur, F, Myles, D. | | Deposit date: | 2013-07-11 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | NEUTRON DIFFRACTION (2.19 Å) | | Cite: | Neutron structure of the cyclic glucose-bound xylose isomerase E186Q mutant.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4K9F
 
 | | Neutron structure of Perdeuterated Rubredoxin refined against 1.75 resolution data collected on the new IMAGINE instrument at HFIR, ORNL | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Munshi, P, Meilleur, F, Myles, D. | | Deposit date: | 2013-04-19 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | NEUTRON DIFFRACTION (1.75 Å) | | Cite: | The IMAGINE instrument: first neutron protein structure and new capabilities for neutron macromolecular crystallography.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3KYX
 
 | |
7T5C
 
 | |
7T5E
 
 | |
