6H3G
 
 | | Alcohol oxidase from Phanerochaete chrysosporium | | Descriptor: | Alcohol oxidase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Nguyen, Q.-T, Romero, E, Dijkman, W.P, de Vasconcellos, S.P, Binda, C, Mattevi, A, Fraaije, M.W. | | Deposit date: | 2018-07-18 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Engineering of Phanerochaete chrysosporium Alcohol Oxidase for Enhanced Oxidative Power toward Glycerol.
Biochemistry, 57, 2018
|
|
5FXD
 
 | | Crystal structure of eugenol oxidase in complex with isoeugenol | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ISOEUGENOL, ... | | Authors: | Nguyen, Q.-T, de Gonzalo, G, Binda, C, Martinez, A.R, Mattevi, A, Fraaije, M.W. | | Deposit date: | 2016-03-01 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biocatalytic Properties and Structural Analysis of Eugenol Oxidase from Rhodococcus Jostii Rha1: A Versatile Oxidative Biocatalyst.
Chembiochem, 17, 2016
|
|
5LXE
 
 | | F420-dependent glucose-6-phosphate dehydrogenase from Rhodococcus jostii RHA1 | | Descriptor: | F420-dependent glucose-6-phosphate dehydrogenase 1, GLYCEROL, SULFATE ION | | Authors: | Nguyen, Q.-T, Trinco, G, Binda, C, Mattevi, A, Fraaije, M.W. | | Deposit date: | 2016-09-20 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery and characterization of an F420-dependent glucose-6-phosphate dehydrogenase (Rh-FGD1) from Rhodococcus jostii RHA1.
Appl. Microbiol. Biotechnol., 101, 2017
|
|
6F97
 
 | | Crystal structure of the V465T mutant of 5-(Hydroxymethyl)furfural Oxidase (HMFO) | | Descriptor: | 5-(hydroxymethyl)furfural oxidase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Pickl, M, Swoboda, A, Romero, E, Winkler, C.K, Binda, C, Mattevi, A, Faber, K, Fraaije, M.W. | | Deposit date: | 2017-12-14 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Kinetic Resolution of sec-Thiols by Enantioselective Oxidation with Rationally Engineered 5-(Hydroxymethyl)furfural Oxidase.
Angew. Chem. Int. Ed. Engl., 57, 2018
|
|
6H0P
 
 | |
2JBR
 
 | |
2JBS
 
 | |
2JBT
 
 | |
5MLN
 
 | |
5L3F
 
 | | LSD1-CoREST1 in complex with polymyxin B | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, Polmyxin B, ... | | Authors: | Speranzini, V, Rotili, D, Ciossani, G, Pilotto, S, Forgione, M, Lucidi, A, Forneris, F, Velankar, S, Mai, A, Mattevi, A. | | Deposit date: | 2016-04-10 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Polymyxins and quinazolines are LSD1/KDM1A inhibitors with unusual structural features.
Sci Adv, 2, 2016
|
|
5LBQ
 
 | | LSD1-CoREST1 in complex with quinazoline-derivative reversible inhibitor | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, N2-(3-(dimethylamino)propyl)-6,7-dimethoxy-N4,N4-dimethylquinazoline-2,4-diamine, ... | | Authors: | Speranzini, V, Rotili, D, Ciossani, G, Pilotto, S, Forgione, M, Lucidi, A, Forneris, F, Velankar, S, Mai, A, Mattevi, A. | | Deposit date: | 2016-06-16 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Polymyxins and quinazolines are LSD1/KDM1A inhibitors with unusual structural features.
Sci Adv, 2, 2016
|
|
6H0N
 
 | |
6HHE
 
 | | Crystal structure of the medfly Odorant Binding Protein CcapOBP22/CcapOBP69a | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, Odorant binding protein OBP69a, SULFATE ION | | Authors: | Falchetto, M, Ciossani, G, Nenci, S, Mattevi, A, Gasperi, G, Forneris, F. | | Deposit date: | 2018-08-28 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.516 Å) | | Cite: | Structural and biochemical evaluation of Ceratitis capitata odorant-binding protein 22 affinity for odorants involved in intersex communication.
Insect Mol.Biol., 28, 2019
|
|
1S3E
 
 | | Crystal structure of MAOB in complex with 6-hydroxy-N-propargyl-1(R)-aminoindan | | Descriptor: | (3R)-3-(PROP-2-YNYLAMINO)INDAN-5-OL, Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Binda, C, Hubalek, F, Li, M, Herzig, Y, Sterling, J, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2004-01-13 | | Release date: | 2004-03-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Monoamine Oxidase B in Complex with Four Inhibitors of the N-Propargylaminoindan Class.
J.Med.Chem., 47, 2004
|
|
1S3B
 
 | | Crystal structure of MAOB in complex with N-methyl-N-propargyl-1(R)-aminoindan | | Descriptor: | Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE, N-[(1S)-2,3-DIHYDRO-1H-INDEN-1-YL]-N-METHYL-N-PROP-2-YNYLAMINE | | Authors: | Binda, C, Hubalek, F, Li, M, Herzig, Y, Sterling, J, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2004-01-13 | | Release date: | 2004-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structures of Monoamine Oxidase B in Complex with Four Inhibitors of the N-Propargylaminoindan Class.
J.Med.Chem., 47, 2004
|
|
1S2Y
 
 | | Crystal structure of MAOB in complex with N-propargyl-1(S)-aminoindan | | Descriptor: | Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE, N-PROPARGYL-1(S)-AMINOINDAN | | Authors: | Binda, C, Hubalek, F, Li, M, Herzig, Y, Sterling, J, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2004-01-12 | | Release date: | 2004-03-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal Structures of Monoamine Oxidase B in Complex with Four Inhibitors of the N-Propargylaminoindan Class.
J.Med.Chem., 47, 2004
|
|
1S2Q
 
 | | Crystal structure of MAOB in complex with N-propargyl-1(R)-aminoindan (Rasagiline) | | Descriptor: | (1R)-N-(prop-2-en-1-yl)-2,3-dihydro-1H-inden-1-amine, Amine oxidase [flavin-containing] B, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Binda, C, Hubalek, F, Li, M, Herzig, Y, Sterling, J, Edmondson, D.E, Mattevi, A. | | Deposit date: | 2004-01-09 | | Release date: | 2004-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal Structures of Monoamine Oxidase B in Complex with Four Inhibitors of the N-Propargylaminoindan Class.
J.Med.Chem., 47, 2004
|
|
5L3E
 
 | | LSD1-CoREST1 in complex with quinazoline-derivative reversible inhibitor | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, N~4~-(1-benzylpiperidin-4-yl)-N~2~-[3-(dimethylamino)propyl]-6,7-dimethoxyquinazoline-2,4-diamine, ... | | Authors: | Speranzini, V, Rotili, D, Ciossani, G, Pilotto, S, Forgione, M, Lucidi, A, Forneris, F, Velankar, S, Mai, A, Mattevi, A. | | Deposit date: | 2016-04-10 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Polymyxins and quinazolines are LSD1/KDM1A inhibitors with unusual structural features.
Sci Adv, 2, 2016
|
|
5L3G
 
 | | LSD1-CoREST1 in complex with polymyxin E (colistin) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, REST corepressor 1, ... | | Authors: | Speranzini, V, Rotili, D, Ciossani, G, Pilotto, S, Forgione, M, Lucidi, A, Forneris, F, Velankar, S, Mai, A, Mattevi, A. | | Deposit date: | 2016-04-10 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Polymyxins and quinazolines are LSD1/KDM1A inhibitors with unusual structural features.
Sci Adv, 2, 2016
|
|
5MXU
 
 | | Structure of the Y503F mutant of vanillyl alcohol oxidase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Vanillyl-alcohol oxidase | | Authors: | Ewing, T.A, Nguyen, Q.-T, Allan, R.C, Gygli, G, Romero, E, Binda, C, Fraaije, M.W, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2017-01-24 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Two tyrosine residues, Tyr-108 and Tyr-503, are responsible for the deprotonation of phenolic substrates in vanillyl-alcohol oxidase.
J. Biol. Chem., 292, 2017
|
|
5MXJ
 
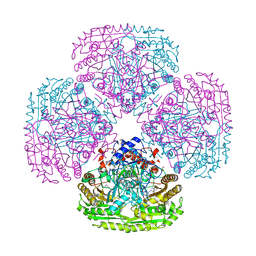 | | Structure of the Y108F mutant of vanillyl alcohol oxidase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Vanillyl-alcohol oxidase | | Authors: | Ewing, T.A, Nguyen, Q.-T, Allan, R.C, Gygli, G, Romero, E, Binda, C, Fraaije, M.W, Mattevi, A, van Berkel, W.J.H. | | Deposit date: | 2017-01-23 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Two tyrosine residues, Tyr-108 and Tyr-503, are responsible for the deprotonation of phenolic substrates in vanillyl-alcohol oxidase.
J. Biol. Chem., 292, 2017
|
|
6SEM
 
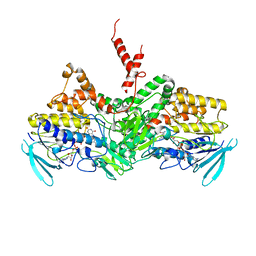 | | Crystal Structure of Ancestral Flavin-containing monooxygenase (FMO) 2 | | Descriptor: | Ancestral Flavin-containing monooxygenase (FMO) 2, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Nicoll, C, Bailleul, G, Fiorentini, F, Mascotti, M.L, Fraaije, M, Mattevi, A. | | Deposit date: | 2019-07-30 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ancestral-sequence reconstruction unveils the structural basis of function in mammalian FMOs.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6SEK
 
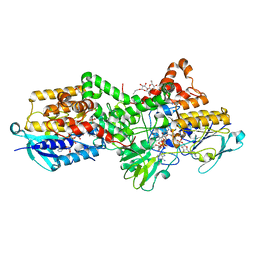 | | Crystal Structure of Ancestral Flavin-containing monooxygenase (FMO) 5 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Ancestral Flavin-containing monooxygenase 5, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Nicoll, C, Bailleul, G, Fiorentini, F, Mascotti, M.L, Fraaije, M, Mattevi, A. | | Deposit date: | 2019-07-30 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ancestral-sequence reconstruction unveils the structural basis of function in mammalian FMOs.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6R1T
 
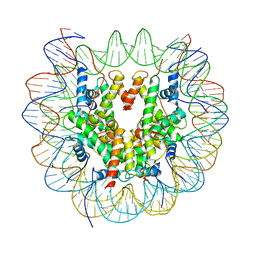 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 1, free nuclesome | | Descriptor: | DNA (147-MER), HISTONE H2A, Histone H2A, ... | | Authors: | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
6R1U
 
 | | Structure of LSD2/NPAC-linker/nucleosome core particle complex: Class 2 | | Descriptor: | DNA (147-MER), FLAVIN-ADENINE DINUCLEOTIDE, Histone H2A, ... | | Authors: | Marabelli, C, Pilotto, S, Chittori, S, Subramaniam, S, Mattevi, A. | | Deposit date: | 2019-03-15 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.
Cell Rep, 27, 2019
|
|
