1OGQ
 
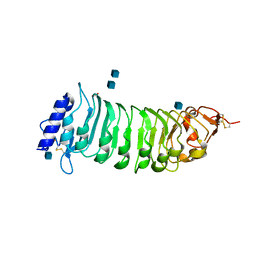 | | The crystal structure of PGIP (polygalacturonase inhibiting protein), a leucine rich repeat protein involved in plant defense | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, POLYGALACTURONASE INHIBITING PROTEIN | | Authors: | Di Matteo, A, Federici, L, Mattei, B, Salvi, G, Johnson, K.A, Savino, C, De Lorenzo, G, Tsernoglou, D, Cervone, F. | | Deposit date: | 2003-05-08 | | Release date: | 2003-07-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Polygalacturonase-Inhibiting Protein (Pgip), a Leucine-Rich Repeat Protein Involved in Plant Defense
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
4UV8
 
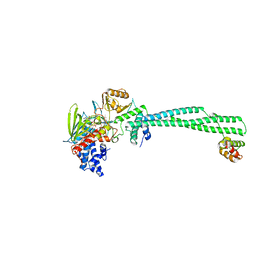 | | LSD1(KDM1A)-CoREST in complex with 1-Benzyl-Tranylcypromine | | Descriptor: | LYSINE-SPECIFIC HISTONE DEMETHYLASE 1A, REST COREPRESSOR 1, [[(2R,3S,4S)-5-[(4aS,10aS)-4a-[(1S)-3-azanylidene-1,4-diphenyl-butyl]-7,8-dimethyl-2,4-bis(oxidanylidene)-5,10a-dihydro-1H-benzo[g]pteridin-10-yl]-2,3,4-tris(oxidanyl)pentoxy]-oxidanyl-phosphoryl] [(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Vianello, P, Botrugno, O, Cappa, A, Ciossani, G, Dessanti, P, Mai, A, Mattevi, A, Meroni, G, Minucci, S, Thaler, F, Tortorici, M, Trifiro, P, Valente, S, Villa, M, Varasi, M, Mercurio, C. | | Deposit date: | 2014-08-05 | | Release date: | 2014-09-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Synthesis, Biological Activity and Mechanistic Insights of 1-Substituted Cyclopropylamine Derivatives: A Novel Class of Irreversible Inhibitors of Histone Demethylase Kdm1A.
Eur.J.Med.Chem., 86C, 2014
|
|
6AFK
 
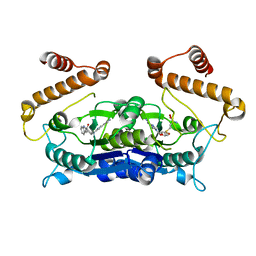 | | Crystal structure of TrmD from Pseudomonas aeruginosa in complex with active-site inhibitor | | Descriptor: | N-{(3S)-1-[3-(pyridin-4-yl)-1H-pyrazol-5-yl]piperidin-3-yl}-1H-indole-2-carboxamide, S-ADENOSYLMETHIONINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Zhong, W, Koay, A, Wong, Y.W, Sahili, A.E, Nah, Q, Kang, C, Poulsen, A, Chionh, Y.K, McBee, M, Matter, A, Hill, J, Lescar, J, Dedon, P.C. | | Deposit date: | 2018-08-08 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Targeting the Bacterial Epitranscriptome for Antibiotic Development: Discovery of Novel tRNA-(N1G37) Methyltransferase (TrmD) Inhibitors.
Acs Infect Dis., 5, 2019
|
|
9FWG
 
 | | LSD1/CoREST bound to bomedemstat | | Descriptor: | Bomedemstat FAD adduct, Lysine-specific histone demethylase 1A, REST corepressor 1 | | Authors: | Speranzini, V, Mattevi, A. | | Deposit date: | 2024-06-30 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Characterization of structural, biochemical, pharmacokinetic, and pharmacodynamic properties of the LSD1 inhibitor bomedemstat in preclinical models.
Prostate, 84, 2024
|
|
8QNR
 
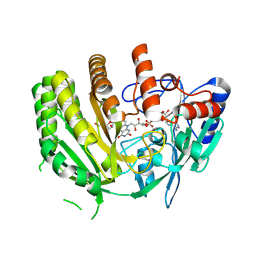 | | Crystal structure of ancestral L-galactono-1,4-lactone dehydrogenase: G413N variant in complex with L-gulono-1,4-lactone | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-galactono-1,4-lactone dehydrogenase, L-gulono-1,4-lactone | | Authors: | Boverio, A, Mattevi, A. | | Deposit date: | 2023-09-27 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of ancestral L-galactono-1,4-lactone dehydrogenase: G413N variant in complex with L-gulono-1,4-lactone
To Be Published
|
|
8QNB
 
 | | Crystal structure of ancestral L-galactono-1,4-lactone dehydrogenase: in complex with L-galactono-1,4-lactone | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-galactono-1,4-lactone, L-galactono-1,4-lactone dehydrogenase | | Authors: | Boverio, A, Mattevi, A. | | Deposit date: | 2023-09-26 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of ancestral L-galactono-1,4-lactone dehydrogenase: in complex with L-galactono-1,4-lactone
To Be Published
|
|
8QNC
 
 | |
8QMY
 
 | |
7AL4
 
 | | Ancestral Flavin-containing monooxygenase (FMO) 1 (mammalian) | | Descriptor: | Ancestral Flavin-containing monooxygenase 1 (mammalian), CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Nicoll, C.R, Mattevi, A. | | Deposit date: | 2020-10-05 | | Release date: | 2020-12-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Ancestral reconstruction of mammalian FMO1 enables structural determination, revealing unique features that explain its catalytic properties.
J.Biol.Chem., 296, 2020
|
|
7O1X
 
 | |
7O2G
 
 | |
7O1R
 
 | |
8Q1G
 
 | | LSD1-CoREST bound to Acetylated K14 of Histone H3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.3C, Lysine-specific histone demethylase 1A, ... | | Authors: | Barone, M, Mattevi, A. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Uncoupling histone modification crosstalk by engineering lysine demethylase LSD1.
Nat.Chem.Biol., 2024
|
|
8Q1H
 
 | | LSD1 Y391K-CoREST bound to Histone H3 N-terminal tail | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.3C, Lysine-specific histone demethylase 1A, ... | | Authors: | Barone, M, Mattevi, A. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Uncoupling histone modification crosstalk by engineering lysine demethylase LSD1.
Nat.Chem.Biol., 2024
|
|
8Q1J
 
 | | LSD1 Y391K-CoREST bound to Acetylated K14 of Histone H3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Histone H3.3C, Lysine-specific histone demethylase 1A, ... | | Authors: | Barone, M, Mattevi, A. | | Deposit date: | 2023-07-31 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Uncoupling histone modification crosstalk by engineering lysine demethylase LSD1.
Nat.Chem.Biol., 2024
|
|
7YWU
 
 | | Eugenol oxidase from rhodococcus jostii: mutant S81H, A423M, H434Y, S394V, I445D, S518P | | Descriptor: | 2-methoxy-4-(prop-2-en-1-yl)phenol, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Alvigini, L, Mattevi, A. | | Deposit date: | 2022-02-14 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure- and computational-aided engineering of an oxidase to produce isoeugenol from a lignin-derived compound.
Nat Commun, 13, 2022
|
|
7YWV
 
 | | Eugenol oxidase from rhodococcus jostii: mutant S81H, D151E, A423M, H434Y, S394V, Q425S, I445D, S518P | | Descriptor: | 2-methoxy-4-(prop-2-en-1-yl)phenol, CALCIUM ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Alvigini, L, Mattevi, A. | | Deposit date: | 2022-02-14 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure- and computational-aided engineering of an oxidase to produce isoeugenol from a lignin-derived compound.
Nat Commun, 13, 2022
|
|
7O2D
 
 | |
7O1Z
 
 | |
5L3D
 
 | | Human LSD1/CoREST: LSD1 Y761H mutation | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, REST corepressor 1 | | Authors: | Pilotto, S, Speranzini, V, Marabelli, C, Mattevi, A. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | LSD1/KDM1A mutations associated to a newly described form of intellectual disability impair demethylase activity and binding to transcription factors.
Hum.Mol.Genet., 25, 2016
|
|
5L3C
 
 | | Human LSD1/CoREST: LSD1 E379K mutation | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Lysine-specific histone demethylase 1A, REST corepressor 1 | | Authors: | Pilotto, S, Speranzini, V, Marabelli, C, Mattevi, A. | | Deposit date: | 2016-04-06 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | LSD1/KDM1A mutations associated to a newly described form of intellectual disability impair demethylase activity and binding to transcription factors.
Hum.Mol.Genet., 25, 2016
|
|
7ZRY
 
 | |
6HD1
 
 | | human STEAP4 bound to NADPH, FAD and heme. | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Oosterheert, W, van Bezouwen, L.S, Rodenburg, R.N.P, Forster, F, Mattevi, A, Gros, P. | | Deposit date: | 2018-08-17 | | Release date: | 2018-10-24 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of human STEAP4 reveal mechanism of iron(III) reduction.
Nat Commun, 9, 2018
|
|
5LXE
 
 | | F420-dependent glucose-6-phosphate dehydrogenase from Rhodococcus jostii RHA1 | | Descriptor: | F420-dependent glucose-6-phosphate dehydrogenase 1, GLYCEROL, SULFATE ION | | Authors: | Nguyen, Q.-T, Trinco, G, Binda, C, Mattevi, A, Fraaije, M.W. | | Deposit date: | 2016-09-20 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Discovery and characterization of an F420-dependent glucose-6-phosphate dehydrogenase (Rh-FGD1) from Rhodococcus jostii RHA1.
Appl. Microbiol. Biotechnol., 101, 2017
|
|
6GZF
 
 | | Xi Class GST from Natrialba magadii | | Descriptor: | Glutathione S-transferase, SULFATE ION | | Authors: | Di Matteo, A, Federici, L, Masulli, M, Carletti, E, Cassidy, J, Paradisi, F, Di Ilio, C, Allocati, N. | | Deposit date: | 2018-07-04 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.614 Å) | | Cite: | Structural Characterization of the Xi Class Glutathione Transferase From the Haloalkaliphilic ArchaeonNatrialba magadii.
Front Microbiol, 10, 2019
|
|
