4YSU
 
 | | Structure of copper nitrite reductase from Geobacillus thermodenitrificans - 25.0 MGy | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, Nitrite reductase, ... | | Authors: | Fukuda, Y, Tse, K.M, Suzuki, M, Diederichs, K, Hirata, K, Nakane, T, Sugahara, M, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Matsumura, H, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-03-17 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Redox-coupled structural changes in nitrite reductase revealed by serial femtosecond and microfocus crystallography
J.Biochem., 159, 2016
|
|
2E2G
 
 | | Crystal structure of archaeal peroxiredoxin, thioredoxin peroxidase from Aeropyrum pernix K1 (pre-oxidation form) | | Descriptor: | Probable peroxiredoxin | | Authors: | Nakamura, T, Yamamoto, T, Abe, M, Matsumura, H, Hagihara, Y, Goto, T, Yamaguchi, T, Inoue, T. | | Deposit date: | 2006-11-13 | | Release date: | 2007-11-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Oxidation of archaeal peroxiredoxin involves a hypervalent sulfur intermediate
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2E2M
 
 | | Crystal structure of archaeal peroxiredoxin, thioredoxin peroxidase from Aeropyrum pernix K1 (sulfinic acid form) | | Descriptor: | Probable peroxiredoxin | | Authors: | Nakamura, T, Yamamoto, T, Abe, M, Matsumura, H, Hagihara, Y, Goto, T, Yamaguchi, T, Inoue, T. | | Deposit date: | 2006-11-14 | | Release date: | 2007-11-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Oxidation of archaeal peroxiredoxin involves a hypervalent sulfur intermediate
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2E4L
 
 | | Thermodynamic and Structural Analysis of Thermolabile RNase HI from Shewanella oneidensis MR-1 | | Descriptor: | Ribonuclease HI | | Authors: | Tadokoro, T, You, D.J, Chon, H, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2006-12-13 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural, thermodynamic, and mutational analyses of a psychrotrophic RNase HI.
Biochemistry, 46, 2007
|
|
1WS8
 
 | | Crystal Structure of Mavicyanin from Cucurbita pepo medullosa (Zucchini) | | Descriptor: | COPPER (II) ION, GLYCEROL, mavicyanin | | Authors: | Xie, Y, Inoue, T, Miyamoto, Y, Matsumura, H, Kunishige, K, Yamaguchi, K, Nojini, M, Suzuki, S, Kai, Y. | | Deposit date: | 2004-11-02 | | Release date: | 2004-11-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural reorganization of the copper binding site involving Thr15 of mavicyanin from Cucurbita pepo medullosa (zucchini) upon reduction.
J.Biochem.(Tokyo), 137, 2005
|
|
1WS7
 
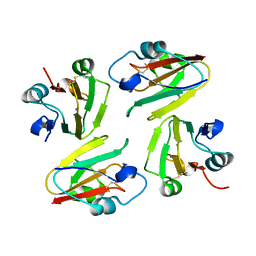 | | Crystal Structure of Mavicyanin from Cucurbita pepo medullosa (Zucchini) | | Descriptor: | COPPER (I) ION, Mavicyanin | | Authors: | Xie, Y, Inoue, T, Miyamoto, Y, Matsumura, H, Kataoka, K, Yamaguchi, K, Nojini, M, Suzuki, S, Kai, Y. | | Deposit date: | 2004-11-02 | | Release date: | 2004-11-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural reorganization of the copper binding site involving Thr15 of mavicyanin from Cucurbita pepo medullosa (zucchini) upon reduction.
J.Biochem.(Tokyo), 137, 2005
|
|
1YC9
 
 | | The crystal structure of the outer membrane protein VceC from the bacterial pathogen Vibrio cholerae at 1.8 resolution | | Descriptor: | MERCURY (II) ION, multidrug resistance protein, octyl beta-D-glucopyranoside | | Authors: | Federici, L, Du, D, Walas, F, Matsumura, H, Fernandez-Recio, J, McKeegan, K.S, Borges-Walmsley, M.I, Luisi, B.F, Walmsley, A.R. | | Deposit date: | 2004-12-22 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the outer membrane protein VCEC from the bacterial pathogen vibrio cholerae at 1.8 A resolution
J.Biol.Chem., 280, 2005
|
|
2CZR
 
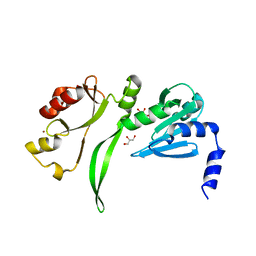 | | Crystal structure of TBP-interacting protein (Tk-TIP26) and implications for its inhibition mechanism of the interaction between TBP and TATA-DNA | | Descriptor: | GLYCEROL, TBP-interacting protein, ZINC ION | | Authors: | Yamamoto, T, Matsuda, T, Inoue, T, Matsumura, H, Morikawa, M, Kanaya, S, Kai, Y. | | Deposit date: | 2005-07-15 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of TBP-interacting protein (Tk-TIP26) and implications for its inhibition mechanism of the interaction between TBP and TATA-DNA
Protein Sci., 15, 2006
|
|
3VKW
 
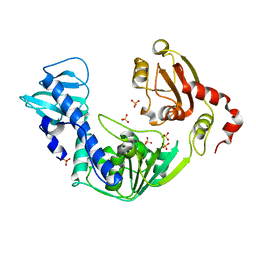 | | Crystal Structure of the Superfamily 1 Helicase from Tomato Mosaic Virus | | Descriptor: | Replicase large subunit, SULFATE ION | | Authors: | Nishikiori, M, Sugiyama, S, Xiang, H, Niiyama, M, Ishibashi, K, Inoue, T, Ishikawa, M, Matsumura, H, Katoh, E. | | Deposit date: | 2011-11-22 | | Release date: | 2012-07-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the superfamily 1 helicase from tomato mosaic virus
J.Virol., 86, 2012
|
|
3VND
 
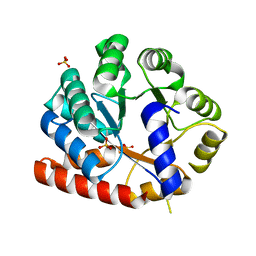 | | Crystal structure of tryptophan synthase alpha-subunit from the psychrophile Shewanella frigidimarina K14-2 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, SULFATE ION, Tryptophan synthase alpha chain | | Authors: | Mitsuya, D, Tanaka, S, Matsumura, H, Takano, K, Urano, N, Ishida, M. | | Deposit date: | 2012-01-12 | | Release date: | 2013-01-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Strategy for cold adaptation of the tryptophan synthase alpha subunit from the psychrophile Shewanella frigidimarina K14-2: crystal structure and physicochemical properties
J.Biochem., 155, 2014
|
|
3WR7
 
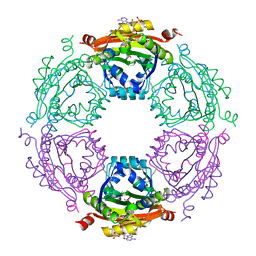 | | Crystal Structure of Spermidine Acetyltransferase from Escherichia coli | | Descriptor: | COENZYME A, SPERMIDINE, Spermidine N1-acetyltransferase | | Authors: | Sugiyama, S, Ishikawa, S, Tomitori, S, Niiyama, M, Hirose, M, Miyazaki, Y, Higashi, K, Adachi, H, Takano, K, Murakami, S, Inoue, T, Mori, Y, Kashiwagi, K, Igarashi, K, Matsumura, H. | | Deposit date: | 2014-02-20 | | Release date: | 2015-09-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism underlying promiscuous polyamine recognition by spermidine acetyltransferase
Int.J.Biochem.Cell Biol., 76, 2016
|
|
3WYQ
 
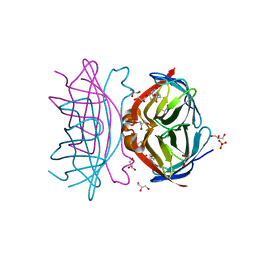 | | Crystal structure of the low-immunogenic core streptavidin mutant LISA-314 (Y22S/Y83S/R84K/E101D/R103K/E116N) at 1.0 A resolution | | Descriptor: | BIOTIN, GLYCEROL, SULFATE ION, ... | | Authors: | Kawato, T, Mizohata, E, Meshizuka, T, Doi, H, Kawamura, T, Matsumura, H, Yumura, K, Tsumoto, K, Kodama, T, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-09-05 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Crystal structure of streptavidin mutant with low immunogenicity.
J.Biosci.Bioeng., 119, 2015
|
|
3WYP
 
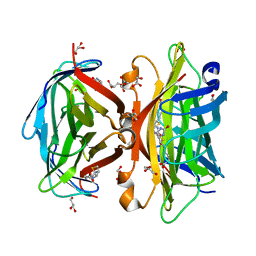 | | Crystal structure of wild-type core streptavidin in complex with D-biotin/biotin-D-sulfoxide at 1.3 A resolution | | Descriptor: | BIOTIN, BIOTIN-D-SULFOXIDE, GLYCEROL, ... | | Authors: | Kawato, T, Mizohata, E, Meshizuka, T, Doi, H, Kawamura, T, Matsumura, H, Yumura, K, Tsumoto, K, Kodama, T, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-09-05 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of streptavidin mutant with low immunogenicity.
J.Biosci.Bioeng., 119, 2015
|
|
3WZQ
 
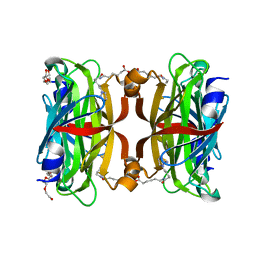 | | Crystal structure of the core streptavidin mutant V212 (Y22S/N23D/S27D/S45N/Y83S/R84K/E101D/R103K/E116N) complexed with iminobiotin long tail (IMNtail) at 1.7 A resolution | | Descriptor: | 6-({5-[(2E,3aS,4S,6aR)-2-iminohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, HEXAETHYLENE GLYCOL, Streptavidin | | Authors: | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Tsumoto, K, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-10-01 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-based design of a streptavidin mutant specific for an artificial biotin analogue.
J.Biochem., 157, 2015
|
|
3WZP
 
 | | Crystal structure of the core streptavidin mutant V21 (Y22S/N23D/S27D/Y83S/R84K/E101D/R103K/E116N) complexed with iminobiotin long tail (IMNtail) at 1.2 A resolution | | Descriptor: | 6-({5-[(2E,3aS,4S,6aR)-2-iminohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, GLYCEROL, Streptavidin | | Authors: | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Tsumoto, K, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-10-01 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure-based design of a streptavidin mutant specific for an artificial biotin analogue.
J.Biochem., 157, 2015
|
|
3X00
 
 | | Crystal structure of the core streptavidin mutant V212 (Y22S/N23D/S27D/S45N/Y83S/R84K/E101D/R103K/E116N) complexed with bis iminobiotin long tail (Bis-IMNtail) at 1.3 A resolution | | Descriptor: | 6-({5-[(2E,3aS,4S,6aR)-2-iminohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, ETHANE-1,2-DIAMINE, Streptavidin | | Authors: | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-10-09 | | Release date: | 2015-01-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure-based design and synthesis of a bivalent iminobiotin analog showing strong affinity toward a low immunogenic streptavidin mutant.
Biosci.Biotechnol.Biochem., 79, 2015
|
|
3WZO
 
 | | Crystal structure of the core streptavidin mutant V21 (Y22S/N23D/S27D/Y83S/R84K/E101D/R103K/E116N) complexed with biotin long tail (BTNtail) at 1.5 A resolution | | Descriptor: | 6-({5-[(3aS,4S,5S,6aR)-5-oxido-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoyl}amino)hexanoic acid, CADMIUM ION, GLYCEROL, ... | | Authors: | Kawato, T, Mizohata, E, Shimizu, Y, Meshizuka, T, Yamamoto, T, Takasu, N, Matsuoka, M, Matsumura, H, Tsumoto, K, Kodama, T, Kanai, M, Doi, H, Inoue, T, Sugiyama, A. | | Deposit date: | 2014-10-01 | | Release date: | 2015-02-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based design of a streptavidin mutant specific for an artificial biotin analogue.
J.Biochem., 157, 2015
|
|
3VGW
 
 | | Crystal structure of monoAc-biotin-avidin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(3aS,4S,6aR)-1-acetyl-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoic acid, Avidin, ... | | Authors: | Terai, T, Maki, E, Sugiyama, S, Takahashi, Y, Matsumura, H, Mori, Y, Nagano, T. | | Deposit date: | 2011-08-21 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rational development of caged-biotin protein-labeling agents and some applications in live cells
Chem.Biol., 18, 2011
|
|
2ZWO
 
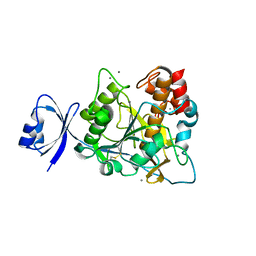 | | Crystal structure of Ca2 site mutant of Pro-S324A | | Descriptor: | CALCIUM ION, Tk-subtilisin | | Authors: | Takeuchi, Y, Tanaka, S, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2008-12-17 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Requirement of a unique Ca(2+)-binding loop for folding of Tk-subtilisin from a hyperthermophilic archaeon.
Biochemistry, 48, 2009
|
|
3VHI
 
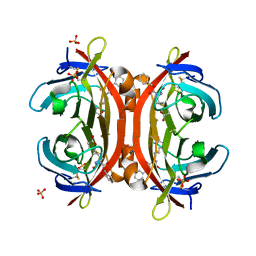 | | Crystal structure of monoZ-biotin-avidin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-{(3aS,4S,6aR)-1-[(benzyloxy)carbonyl]-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl}pentanoic acid, Avidin, ... | | Authors: | Terai, T, Maki, E, Sugiyama, S, Takahashi, Y, Matsumura, H, Mori, Y, Nagano, T. | | Deposit date: | 2011-08-25 | | Release date: | 2011-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Rational development of caged-biotin protein-labeling agents and some applications in live cells
Chem.Biol., 18, 2011
|
|
3VI2
 
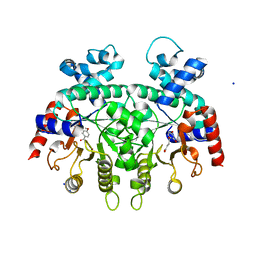 | | Crystal Structure Analysis of Plasmodium falciparum OMP Decarboxylase in complex with inhibitor HMOA | | Descriptor: | 4-(2-hydroxy-4-methoxyphenyl)-4-oxobutanoic acid, Orotidine 5'-phosphate decarboxylase, SODIUM ION | | Authors: | Takashima, Y, Mizohata, E, Krungkrai, S.R, Matsumura, H, Krungkrai, J, Horii, T, Inoue, T. | | Deposit date: | 2011-09-16 | | Release date: | 2012-08-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The in silico screening and X-ray structure analysis of the inhibitor complex of Plasmodium falciparum orotidine 5'-monophosphate decarboxylase
J.Biochem., 152, 2012
|
|
2ZWP
 
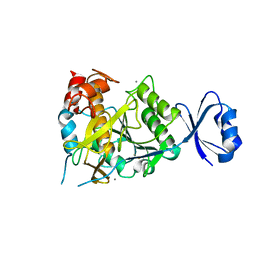 | | Crystal structure of Ca3 site mutant of Pro-S324A | | Descriptor: | CALCIUM ION, Tk-subtilisin | | Authors: | Takeuchi, Y, Tanaka, S, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2008-12-17 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Requirement of a unique Ca(2+)-binding loop for folding of Tk-subtilisin from a hyperthermophilic archaeon.
Biochemistry, 48, 2009
|
|
3VHM
 
 | | Crystal structure of NPC-biotin-avidin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-[(3aS,4R,6aR)-1-{[(1R)-1-(6-nitro-1,3-benzodioxol-5-yl)ethoxy]carbonyl}-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanoic acid, Avidin, ... | | Authors: | Terai, T, Maki, E, Sugiyama, S, Takahashi, Y, Matsumura, H, Mori, Y, Nagano, T. | | Deposit date: | 2011-08-29 | | Release date: | 2011-12-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational development of caged-biotin protein-labeling agents and some applications in live cells
Chem.Biol., 18, 2011
|
|
3W6G
 
 | | Structure of peroxiredoxin from anaerobic hyperthermophilic archaeon Pyrococcus horikoshii | | Descriptor: | CITRATE ANION, Probable peroxiredoxin | | Authors: | Nakamura, T, Mori, A, Niiyama, M, Matsumura, H, Tokuyama, C, Morita, J, Uegaki, K, Inoue, T. | | Deposit date: | 2013-02-14 | | Release date: | 2013-07-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of peroxiredoxin from the anaerobic hyperthermophilic archaeon Pyrococcus horikoshii
Acta Crystallogr.,Sect.F, 69, 2013
|
|
3WQT
 
 | | Staphylococcus aureus FtsA complexed with AMPPNP | | Descriptor: | CHLORIDE ION, Cell division protein FtsA, MAGNESIUM ION, ... | | Authors: | Fujita, J, Maeda, Y, Miyazaki, Y, Inoue, T, Matsumura, H. | | Deposit date: | 2014-02-01 | | Release date: | 2014-10-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of FtsA from Staphylococcus aureus
FEBS Lett., 588, 2014
|
|
