5YE4
 
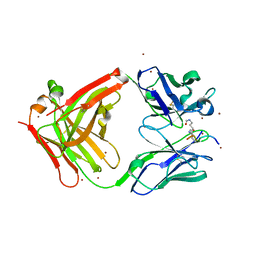 | | Crystal structure of the complex of di-acetylated histone H4 and 1A9D7 Fab fragment | | Descriptor: | 1A9D7 L chain, 1A9D7 VH CH1 chain, ZINC ION, ... | | Authors: | Matsuda, T, Ito, T, Wakamori, M, Umehara, T. | | Deposit date: | 2017-09-15 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | JQ1 affects BRD2-dependent and independent transcription regulation without disrupting H4-hyperacetylated chromatin states.
Epigenetics, 13, 2018
|
|
5YE3
 
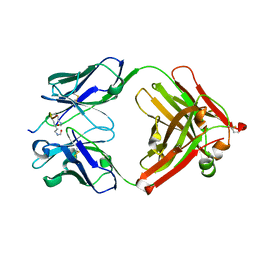 | | Crystal structure of the complex of di-acetylated histone H4 and 2A7D9 Fab fragment | | Descriptor: | 2A7D9 L chain, 2A7D9 VH CH1 chain, di-acetylated histone H4 | | Authors: | Matsuda, T, Ito, T, Wakamori, M, Umehara, T. | | Deposit date: | 2017-09-15 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | JQ1 affects BRD2-dependent and independent transcription regulation without disrupting H4-hyperacetylated chromatin states.
Epigenetics, 13, 2018
|
|
5XWD
 
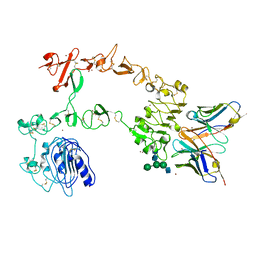 | | Crystal structure of the complex of 059-152-Fv and EGFR-ECD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, ... | | Authors: | Matsuda, T, Ito, T, Shirouzu, M. | | Deposit date: | 2017-06-29 | | Release date: | 2018-02-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Cell-free synthesis of functional antibody fragments to provide a structural basis for antibody-antigen interaction
PLoS ONE, 13, 2018
|
|
3UFZ
 
 | | Crystal structure of a Trp-less green fluorescent protein translated by the universal genetic code | | Descriptor: | Green fluorescent protein | | Authors: | Kawahara-Kobayashi, A, Araiso, Y, Matsuda, T, Yokoyama, S, Kigawa, T, Nureki, O, Kiga, D. | | Deposit date: | 2011-11-02 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Simplification of the genetic code: restricted diversity of genetically encoded amino acids.
Nucleic Acids Res., 40, 2012
|
|
3UG0
 
 | | Crystal structure of a Trp-less green fluorescent protein translated by the simplified genetic code | | Descriptor: | Green fluorescent protein | | Authors: | Kawahara-Kobayashi, A, Araiso, Y, Matsuda, T, Yokoyama, S, Kigawa, T, Nureki, O, Kiga, D. | | Deposit date: | 2011-11-02 | | Release date: | 2012-10-17 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Simplification of the genetic code: restricted diversity of genetically encoded amino acids.
Nucleic Acids Res., 40, 2012
|
|
1V9J
 
 | | Solution structure of a BolA-like protein from Mus musculus | | Descriptor: | BolA-like protein RIKEN cDNA 1110025L05 | | Authors: | Kasai, T, Inoue, M, Koshiba, S, Yabuki, T, Aoki, M, Nunokawa, E, Seki, E, Matsuda, T, Matsuda, N, Tomo, Y, Shirouzu, M, Terada, T, Obayashi, N, Hamana, H, Shinya, N, Tatsuguchi, A, Yasuda, S, Yoshida, M, Hirota, H, Matsuo, Y, Tani, K, Suzuki, H, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-26 | | Release date: | 2004-02-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a BolA-like protein from Mus musculus
Protein Sci., 13, 2004
|
|
2CZR
 
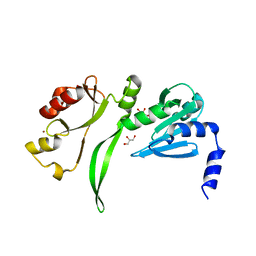 | | Crystal structure of TBP-interacting protein (Tk-TIP26) and implications for its inhibition mechanism of the interaction between TBP and TATA-DNA | | Descriptor: | GLYCEROL, TBP-interacting protein, ZINC ION | | Authors: | Yamamoto, T, Matsuda, T, Inoue, T, Matsumura, H, Morikawa, M, Kanaya, S, Kai, Y. | | Deposit date: | 2005-07-15 | | Release date: | 2006-02-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of TBP-interacting protein (Tk-TIP26) and implications for its inhibition mechanism of the interaction between TBP and TATA-DNA
Protein Sci., 15, 2006
|
|
1IOZ
 
 | | Crystal Structure of the C-HA-RAS Protein Prepared by the Cell-Free Synthesis | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, TRANSFORMING PROTEIN P21/H-RAS-1 | | Authors: | Kigawa, T, Yamaguchi-Nunokawa, E, Kodama, K, Matsuda, T, Yabuki, T, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2001-04-18 | | Release date: | 2001-10-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selenomethionine incorporation into a protein by cell-free synthesis
J.STRUCT.FUNCT.GENOM., 2, 2001
|
|
2UXS
 
 | | 2.7A crystal structure of inorganic pyrophosphatase (Rv3628) from Mycobacterium tuberculosis at pH 7.5 | | Descriptor: | INORGANIC PYROPHOSPHATASE, PHOSPHATE ION | | Authors: | Cole, R.E, Cianci, M, Hall, J.F, Matsuda, T, Kigawa, T, Yokoyama, S, Hasnain, S.S, Tabernero, L. | | Deposit date: | 2007-03-29 | | Release date: | 2008-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Rv3628: An Inorganic Pyrophosphatase from Mycobacterium Tuberculosis
To be Published
|
|
6J1C
 
 | | Photoswitchable fluorescent protein Gamillus, N150C/T204V double mutant, off-state | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein | | Authors: | Nakashima, R, Shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2018-12-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Acid-Tolerant Reversibly Switchable Green Fluorescent Protein for Super-resolution Imaging under Acidic Conditions.
Cell Chem Biol, 26, 2019
|
|
6J1B
 
 | | Photoswitchable fluorescent protein Gamillus, N150C/T204V double mutant, on-state | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Nakashima, R, Shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2018-12-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Acid-Tolerant Reversibly Switchable Green Fluorescent Protein for Super-resolution Imaging under Acidic Conditions.
Cell Chem Biol, 26, 2019
|
|
6J1A
 
 | | Photoswitchable fluorescent protein Gamillus, off-state | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Nakashima, R, Sakurai, K, shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2018-12-28 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Acid-Tolerant Reversibly Switchable Green Fluorescent Protein for Super-resolution Imaging under Acidic Conditions.
Cell Chem Biol, 26, 2019
|
|
6JXF
 
 | | Photoswitchable fluorescent protein Gamillus, off-state (pH7.0) | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Nakashima, R, Sakurai, K, shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2019-04-23 | | Release date: | 2019-11-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Acid-Tolerant Reversibly Switchable Green Fluorescent Protein for Super-resolution Imaging under Acidic Conditions.
Cell Chem Biol, 26, 2019
|
|
3WCK
 
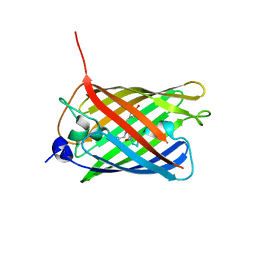 | | Crystal structure of monomeric photosensitizing fluorescent protein, Supernova | | Descriptor: | Monomeric photosenitizing fluorescent protein supernova | | Authors: | Sakai, N, Matsuda, T, Takemoto, K, Nagai, T. | | Deposit date: | 2013-05-27 | | Release date: | 2013-10-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | SuperNova, a monomeric photosensitizing fluorescent protein for chromophore-assisted light inactivation
Sci Rep, 3, 2013
|
|
6ISV
 
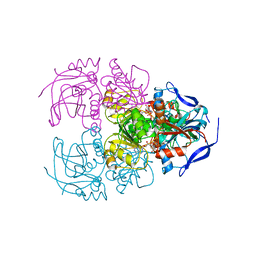 | | Structure of acetophenone reductase from Geotrichum candidum NBRC 4597 in complex with NAD | | Descriptor: | Acetophenone reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | Authors: | Koesoema, A.A, Sugiyama, Y, Senda, M, Senda, T, Matsuda, T. | | Deposit date: | 2018-11-19 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for a highly (S)-enantioselective reductase towards aliphatic ketones with only one carbon difference between side chain.
Appl.Microbiol.Biotechnol., 103, 2019
|
|
3A8S
 
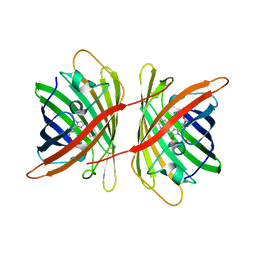 | |
5Y01
 
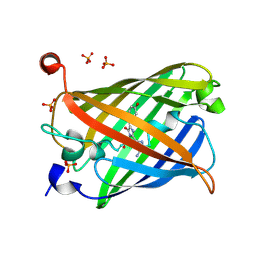 | | Acid-tolerant monomeric GFP, Gamillus, non-fluorescence (OFF) state | | Descriptor: | Green fluorescent protein, PHOSPHATE ION | | Authors: | Nakashima, R, Sakurai, K, Shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2017-07-14 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Acid-Tolerant Monomeric GFP from Olindias formosa.
Cell Chem Biol, 25, 2018
|
|
5Y00
 
 | | Acid-tolerant monomeric GFP, Gamillus, fluorescence (ON) state | | Descriptor: | CHLORIDE ION, GLYCEROL, Green fluorescent protein, ... | | Authors: | Nakashima, R, Sakurai, K, Shinoda, H, Matsuda, T, Nagai, T. | | Deposit date: | 2017-07-14 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Acid-Tolerant Monomeric GFP from Olindias formosa.
Cell Chem Biol, 25, 2018
|
|
2FYH
 
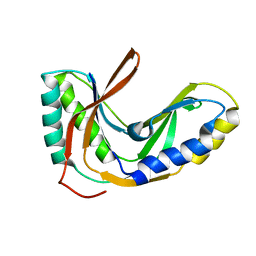 | | Solution structure of the 2'-5' RNA ligase-like protein from Pyrococcus furiosus | | Descriptor: | putative integral membrane transport protein | | Authors: | Okada, K, Matsuda, T, Sakamoto, T, Muto, Y, Yokoyama, S, Kanai, A, Kawai, G, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-02-08 | | Release date: | 2007-02-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Characterization of a heat-stable enzyme possessing GTP-dependent RNA ligase activity from a hyperthermophilic archaeon, Pyrococcus furiosus
Rna, 15, 2009
|
|
1VDY
 
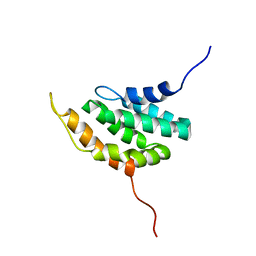 | | NMR Structure of the hypothetical ENTH-VHS domain At3g16270 from Arabidopsis thaliana | | Descriptor: | hypothetical protein (RAFL09-17-B18) | | Authors: | Lopez-Mendez, B, Pantoja-Uceda, D, Tomizawa, T, Koshiba, S, Kigawa, T, Shirouzu, M, Terada, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Seki, M, Shinozaki, K, Yokoyama, S, Guntert, P, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-03-25 | | Release date: | 2005-05-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the hypothetical ENTH-VHS domain AT3G16270 from arabidopsis thaliana
To be Published
|
|
1UFW
 
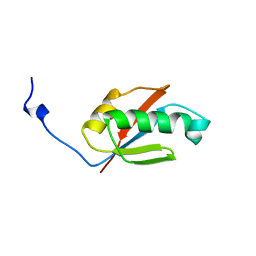 | | Solution structure of RNP domain in Synaptojanin 2 | | Descriptor: | Synaptojanin 2 | | Authors: | He, F, Muto, Y, Ushikoshi, R, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Ohara, O, Nagase, T, Kikuno, R, Nakayama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-10 | | Release date: | 2003-12-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RNP domain in Synaptojanin 2
To be Published
|
|
1UFN
 
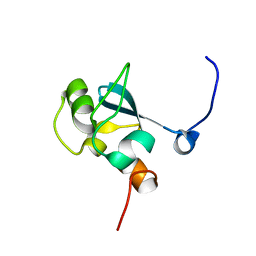 | | Solution structure of the SAND domain of the putative nuclear protein homolog (5830484A20Rik) | | Descriptor: | putative nuclear protein homolog 5830484A20Rik | | Authors: | Tochio, N, Kobayashi, N, Koshiba, S, Kigawa, T, Inoue, M, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Matsuo, Y, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-02 | | Release date: | 2004-06-22 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SAND domain of the putative nuclear protein homolog (5830484A20Rik)
To be Published
|
|
1UGK
 
 | | Solution structure of the first C2 domain of synaptotagmin IV from human fetal brain (KIAA1342) | | Descriptor: | Synaptotagmin IV | | Authors: | Nagashima, T, Hayashi, F, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Matsuo, Y, Ohara, O, Nagase, T, Kikuno, R, Nakayama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-16 | | Release date: | 2003-12-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first C2 domain of synaptotagmin IV from human fetal brain (KIAA1342)
To be Published
|
|
1UGV
 
 | | Solution structure of the SH3 domain of human olygophrein-1 like protein (KIAA0621) | | Descriptor: | Olygophrenin-1 like protein | | Authors: | Inoue, K, Hayashi, F, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Matsuo, Y, Ohara, O, Nagase, T, Kikuno, R, Nakayama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-20 | | Release date: | 2003-12-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of human olygophrein-1 like protein (KIAA0621)
To be Published
|
|
1UG8
 
 | | NMR structure of the R3H domain from Poly(A)-specific Ribonuclease | | Descriptor: | Poly(A)-specific Ribonuclease | | Authors: | Nagata, T, Muto, Y, Hayami, N, Uda, H, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Kobayashi, N, Tanaka, A, Osanai, T, Matsuo, Y, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-15 | | Release date: | 2004-08-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the R3H domain from Poly(A)-specific Ribonuclease
To be Published
|
|
