6HTO
 
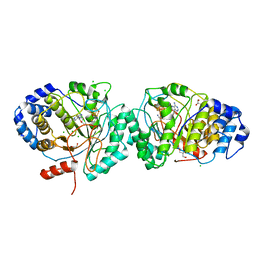 | | Tryptophan lyase 'empty state' | | Descriptor: | 3-methyl-2-indolic acid synthase, 5'-DEOXYADENOSINE, CHLORIDE ION, ... | | Authors: | Amara, P, Mouesca, J.M, Bella, M, Martin, L, Saragaglia, C, Gambarelli, S, Nicolet, Y. | | Deposit date: | 2018-10-04 | | Release date: | 2018-11-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Radical S-Adenosyl-l-methionine Tryptophan Lyase (NosL): How the Protein Controls the Carboxyl Radical •CO2-Migration.
J.Am.Chem.Soc., 140, 2018
|
|
3UAG
 
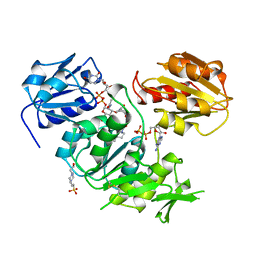 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Bertrand, J.A, Auger, G, Martin, L, Fanchon, E, Blanot, D, Le Beller, D, Van Heijenoort, J, Dideberg, O. | | Deposit date: | 1999-02-24 | | Release date: | 2000-02-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Determination of the MurD mechanism through crystallographic analysis of enzyme complexes.
J.Mol.Biol., 289, 1999
|
|
5FEW
 
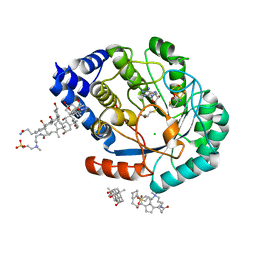 | | HydE from T. maritima in complex with S-adenosyl-L-cysteine (final product) | | Descriptor: | (2R,4R)-2-methyl-1,3-thiazolidine-2,4-dicarboxylic acid, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, ... | | Authors: | Rohac, R, Amara, P, Benjdia, A, Martin, L, Ruffie, P, Favier, A, Berteau, O, Mouesca, J.M, Fontecilla-Camps, J.C, Nicolet, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Carbon-sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE.
Nat.Chem., 8, 2016
|
|
1EEH
 
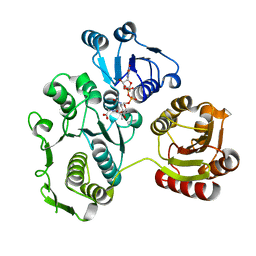 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | Descriptor: | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE, URIDINE-5'-DIPHOSPHATE-N-ACETYLMURAMOYL-L-ALANINE | | Authors: | Bertrand, J.A, Fanchon, E, Martin, L, Chantalat, L, Auger, G, Blanot, D, van Heijenoort, J, Dideberg, O. | | Deposit date: | 2000-01-31 | | Release date: | 2001-01-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | "Open" structures of MurD: domain movements and structural similarities with folylpolyglutamate synthetase.
J.Mol.Biol., 301, 2000
|
|
5FEX
 
 | | HydE from T. maritima in complex with Se-adenosyl-L-selenocysteine (tfinal of the reaction) | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, 5'-DEOXYADENOSINE, CHLORIDE ION, ... | | Authors: | Rohac, R, Amara, P, Benjdia, A, Martin, L, Ruffie, P, Favier, A, Berteau, O, Mouesca, J.M, Fontecilla-Camps, J.C, Nicolet, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Carbon-sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE.
Nat.Chem., 8, 2016
|
|
5MQ1
 
 | | Crystal structure of the BRD7 bromodomain in complex with BI-9564 | | Descriptor: | 1,2-ETHANEDIOL, 4-[4-[(dimethylamino)methyl]-2,5-dimethoxy-phenyl]-2-methyl-2,7-naphthyridin-1-one, Bromodomain-containing protein 7, ... | | Authors: | Diaz-Saez, L, Martin, L.J, Panagakou, I, Picaud, S, Krojer, T, von Delft, F, Knapp, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Huber, K, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-12-19 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the BRD7 bromodomain in complex with BI-9564
To Be Published
|
|
5EU1
 
 | | CRYSTAL STRUCTURE OF BRD9 IN COMPLEX WITH BI-7273 | | Descriptor: | 4-[4-[(dimethylamino)methyl]-3,5-dimethoxy-phenyl]-2-methyl-2,7-naphthyridin-1-one, BRD9 | | Authors: | Bader, G, Martin, L.M, Steurer, S, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2015-11-18 | | Release date: | 2016-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Based Design of an in Vivo Active Selective BRD9 Inhibitor.
J.Med.Chem., 59, 2016
|
|
7PD1
 
 | | Crystal structure of the L-tyrosine-bound radical SAM tyrosine lyase ThiH (2-iminoacetate synthase) from Thermosinus carboxydivorans | | Descriptor: | 5'-DEOXYADENOSINE, BROMIDE ION, GLYCEROL, ... | | Authors: | Amara, P, Saragaglia, C, Mouesca, J.-M, Martin, L, Nicolet, Y. | | Deposit date: | 2021-08-04 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | L-tyrosine-bound ThiH structure reveals C-C bond break differences within radical SAM aromatic amino acid lyases.
Nat Commun, 13, 2022
|
|
5FF3
 
 | | HydE from T. maritima in complex with 4R-TCA | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Rohac, R, Amara, P, Benjdia, A, Martin, L, Ruffie, P, Favier, A, Berteau, O, Mouesca, J.M, Fontecilla-Camps, J.C, Nicolet, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Carbon-sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE.
Nat.Chem., 8, 2016
|
|
5FF0
 
 | | HydE from T. maritima in complex with S-adenosyl-L-cysteine and methionine | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, Fe4-Se4 cluster, ... | | Authors: | Rohac, R, Amara, P, Benjdia, A, Martin, L, Ruffie, P, Favier, A, Berteau, O, Mouesca, J.M, Fontecilla-Camps, J.C, Nicolet, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Carbon-sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE.
Nat.Chem., 8, 2016
|
|
3H3X
 
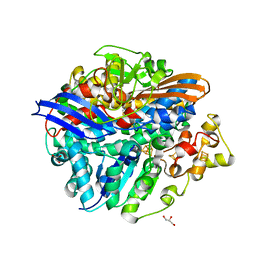 | | Structure of the V74M large subunit mutant of NI-FE hydrogenase in an oxidized state | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martinez, N, Martin, L, Fontecilla-Camps, J.C. | | Deposit date: | 2009-04-17 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Introduction of methionines in the gas channel makes [NiFe] hydrogenase aero-tolerant
J.Am.Chem.Soc., 131, 2009
|
|
7PD2
 
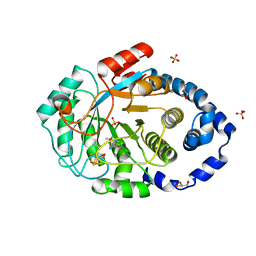 | | Crystal structure of the substrate-free radical SAM tyrosine lyase ThiH (2-iminoacetate synthase) from Thermosinus carboxydivorans | | Descriptor: | 5'-DEOXYADENOSINE, GLYCEROL, IRON/SULFUR CLUSTER, ... | | Authors: | Amara, P, Saragaglia, C, Mouesca, J.-M, Martin, L, Nicolet, Y. | | Deposit date: | 2021-08-04 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | L-tyrosine-bound ThiH structure reveals C-C bond break differences within radical SAM aromatic amino acid lyases.
Nat Commun, 13, 2022
|
|
5FF2
 
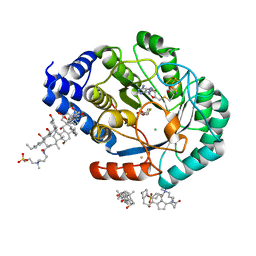 | | HydE from T. maritima in complex with (2R,4R)-TDA | | Descriptor: | (2~{R},4~{R})-1,3-thiazolidine-2,4-dicarboxylic acid, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, ... | | Authors: | Rohac, R, Amara, P, Benjdia, A, Martin, L, Ruffie, P, Favier, A, Berteau, O, Mouesca, J.M, Fontecilla-Camps, J.C, Nicolet, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Carbon-sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE.
Nat.Chem., 8, 2016
|
|
5FF4
 
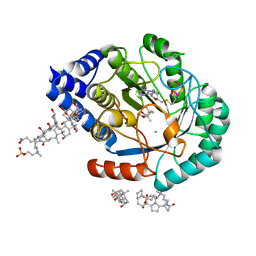 | | HydE from T. maritima in complex with (2R,4R)-TMeTDA | | Descriptor: | (2~{R},4~{R})-2,5,5-trimethyl-1,3-thiazolidine-2,4-dicarboxylic acid, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, ... | | Authors: | Rohac, R, Amara, P, Benjdia, A, Martin, L, Ruffie, P, Favier, A, Berteau, O, Mouesca, J.M, Fontecilla-Camps, J.C, Nicolet, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Carbon-sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE.
Nat.Chem., 8, 2016
|
|
2PZY
 
 | | Structure of MK2 Complexed with Compound 76 | | Descriptor: | (4R)-N-[4-({[2-(DIMETHYLAMINO)ETHYL]AMINO}CARBONYL)-1,3-THIAZOL-2-YL]-4-METHYL-1-OXO-2,3,4,9-TETRAHYDRO-1H-BETA-CARBOLINE-6-CARBOXAMIDE, MAP kinase-activated protein kinase 2, STAUROSPORINE | | Authors: | White, A, Wu, J.P, Wang, J, Abeywardane, A, Andersen, D, Emmanuel, M, Gautschi, E, Goldberg, D.R, Kashem, M.A, Lukas, S, Mao, W, Martin, L, Morwick, T, Moss, N, Pargellis, C, Patel, U.R, Patnaude, L, Peet, G.W, Skow, D, Snow, R.J, Ward, Y, Werneburg, B. | | Deposit date: | 2007-05-18 | | Release date: | 2007-07-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The discovery of carboline analogs as potent MAPKAP-K2 inhibitors
Bioorg.Med.Chem.Lett., 17, 2007
|
|
8QML
 
 | | (2R,4R)-MeTDA bound HydE structure (control experiment) | | Descriptor: | (2R,4R)-2-methyl-1,3-thiazolidine-2,4-dicarboxylic acid, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, ... | | Authors: | Omeiri, J, Martin, L, Usclat, A, Cherrier, M.V, Nicolet, Y. | | Deposit date: | 2023-09-22 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Maturation of the [FeFe]-Hydrogenase: Direct Transfer of the ( kappa 3 -cysteinate)Fe II (CN)(CO) 2 Complex B from HydG to HydE.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
1YQ9
 
 | | Structure of the unready oxidized form of [NiFe] hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Cavazza, C, Matho, M, Faber, B.W, Roseboom, W, Albracht, S.P, Garcin, E, Rousset, M, Fontecilla-Camps, J.C. | | Deposit date: | 2005-02-01 | | Release date: | 2005-04-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural differences between the ready and unready oxidized states of [NiFe] hydrogenases.
J.Biol.Inorg.Chem., 10, 2005
|
|
5FEZ
 
 | | HydE from T. maritima in complex with (2R,4R)-MeSeTDA, 5'-deoxyadenosine and methionine | | Descriptor: | (2~{R},4~{R})-2-methyl-1,3-selenazolidine-2,4-dicarboxylic acid, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, 5'-DEOXYADENOSINE, ... | | Authors: | Rohac, R, Amara, P, Benjdia, A, Martin, L, Ruffie, P, Favier, A, Berteau, O, Mouesca, J.M, Fontecilla-Camps, J.C, Nicolet, Y. | | Deposit date: | 2015-12-17 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Carbon-sulfur bond-forming reaction catalysed by the radical SAM enzyme HydE.
Nat.Chem., 8, 2016
|
|
1ZLQ
 
 | | Crystallographic and spectroscopic evidence for high affinity binding of Fe EDTA (H2O)- to the periplasmic nickel transporter NikA | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, ACETATE ION, CHLORIDE ION, ... | | Authors: | Cherrier, M.V, Martin, L, Cavazza, C, Jacquamet, L, Lemaire, D, Gaillard, J, Fontecilla Camps, J.C. | | Deposit date: | 2005-05-09 | | Release date: | 2005-08-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic and Spectroscopic Evidence for High Affinity Binding of FeEDTA(H(2)O)(-) to the Periplasmic Nickel Transporter NikA
J.Am.Chem.Soc., 127, 2005
|
|
8QMM
 
 | | M291I variant of the [FeFe]-hydrogenase maturase HydE from Thermotoga maritima | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CHLORIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Omeiri, J, Martin, L, Usclat, A, Cherrier, M.V, Nicolet, Y. | | Deposit date: | 2023-09-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Maturation of the [FeFe]-Hydrogenase: Direct Transfer of the ( kappa 3 -cysteinate)Fe II (CN)(CO) 2 Complex B from HydG to HydE.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8QMK
 
 | | Enzymatically-produced complex-B bound TmHydE structure | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, CARBON MONOXIDE, CHLORIDE ION, ... | | Authors: | Omeiri, J, Martin, L, Usclat, A, Cherrier, M.V, Nicolet, Y. | | Deposit date: | 2023-09-22 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Maturation of the [FeFe]-Hydrogenase: Direct Transfer of the ( kappa 3 -cysteinate)Fe II (CN)(CO) 2 Complex B from HydG to HydE.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
5F25
 
 | | Crystal structure of the BRD9 bromodomain in complex with compound 4. | | Descriptor: | 4-(1,5-dimethyl-6-oxidanylidene-pyridin-3-yl)benzamide, BRD9 | | Authors: | Bader, G, Martin, L.J, Steurer, S, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2015-12-01 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure-Based Design of an in Vivo Active Selective BRD9 Inhibitor.
J.Med.Chem., 59, 2016
|
|
5F1H
 
 | | Crystal structure of the BRD9 bromodamian in complex with BI-9564. | | Descriptor: | 4-[4-[(dimethylamino)methyl]-2,5-dimethoxy-phenyl]-2-methyl-2,7-naphthyridin-1-one, Bromodomain-containing protein 9 | | Authors: | Bader, G, Martin, L.J, Steurer, S, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2015-11-30 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-Based Design of an in Vivo Active Selective BRD9 Inhibitor.
J.Med.Chem., 59, 2016
|
|
5F1L
 
 | | Crystal structure of the bromodomain of BRD9 in complex with compound 9. | | Descriptor: | 5-[3,5-dimethoxy-4-[(3-oxidanylazetidin-1-yl)methyl]phenyl]-1,3-dimethyl-pyridin-2-one, Bromodomain-containing protein 9 | | Authors: | Bader, G, Martin, L.J, Steurer, S, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2015-11-30 | | Release date: | 2016-03-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design of an in Vivo Active Selective BRD9 Inhibitor.
J.Med.Chem., 59, 2016
|
|
6FD2
 
 | | Radical SAM 1,2-diol dehydratase AprD4 in complex with its substrate paromamine | | Descriptor: | 5'-DEOXYADENOSINE, IRON/SULFUR CLUSTER, METHIONINE, ... | | Authors: | Liu, W.Q, Amara, P, Mouesca, J.M, Ji, X, Renoux, O, Martin, L, Zhang, C, Zhang, Q, Nicolet, Y. | | Deposit date: | 2017-12-21 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | 1,2-Diol Dehydration by the Radical SAM Enzyme AprD4: A Matter of Proton Circulation and Substrate Flexibility.
J. Am. Chem. Soc., 140, 2018
|
|
