7QXR
 
 | | Azacoelenterazine-bound Renilla-type luciferase (AncFT) | | Descriptor: | 3-(4-hydroxyphenyl)-8-[(4-hydroxyphenyl)methyl]-5-(phenylmethyl)-1$l^{4},4,7,8-tetrazabicyclo[4.3.0]nona-1(6),2,4-trien-9-one, Fragment transplantation onto a hyperstable ancestor of haloalkane dehalogenases and Renilla luciferase (Anc-FT) | | Authors: | Marek, M, Schenkmayerova, A, Janin, Y.L. | | Deposit date: | 2022-01-27 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Catalytic mechanism for Renilla-type luciferases
Nat Catal, 2023
|
|
7QXQ
 
 | | Coelenteramide-bound Renilla-type luciferase (AncFT) | | Descriptor: | Fragment transplantation onto hyperstable ancestor of haloalkane dehalogenases and Renilla luciferase (Anc-FT), N-[3-BENZYL-5-(4-HYDROXYPHENYL)PYRAZIN-2-YL]-2-(4-HYDROXYPHENYL)ACETAMIDE | | Authors: | Marek, M, Schenkmayerova, A. | | Deposit date: | 2022-01-27 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Catalytic mechanism for Renilla-type luciferases
Nat Catal, 2023
|
|
8OE2
 
 | | Structure of hyperstable haloalkane dehalogenase variant DhaA223 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Marek, M. | | Deposit date: | 2023-03-10 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Advancing Enzyme's Stability and Catalytic Efficiency through Synergy of Force-Field Calculations, Evolutionary Analysis, and Machine Learning.
Acs Catalysis, 13, 2023
|
|
8OE6
 
 | | Structure of hyperstable haloalkane dehalogenase variant DhaA231 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Structure of hyperstable haloalkane dehalogenase variant DhaA231 | | Authors: | Marek, M. | | Deposit date: | 2023-03-10 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Advancing Enzyme's Stability and Catalytic Efficiency through Synergy of Force-Field Calculations, Evolutionary Analysis, and Machine Learning.
Acs Catalysis, 13, 2023
|
|
5FUE
 
 | |
8OM6
 
 | |
8BO9
 
 | | NanoLuc-D9R/H57A/K89R mutant complexed with azacoelenterazine bound in intra-barrel catalytic site | | Descriptor: | 3-(4-hydroxyphenyl)-8-[(4-hydroxyphenyl)methyl]-5-(phenylmethyl)-1$l^{4},4,7,8-tetrazabicyclo[4.3.0]nona-1(6),2,4-trien-9-one, Non structural polyprotein | | Authors: | Marek, M, Janin, L.Y. | | Deposit date: | 2022-11-15 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Illuminating the mechanism and allosteric behavior of NanoLuc luciferase.
Nat Commun, 14, 2023
|
|
7NFZ
 
 | |
8CE0
 
 | | N-terminal domain of human apolipoprotein E | | Descriptor: | GLYCEROL, Maltodextrin-binding protein,Apolipoprotein E | | Authors: | Marek, M, Nemergut, M. | | Deposit date: | 2023-02-01 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Domino-like effect of C112R mutation on ApoE4 aggregation and its reduction by Alzheimer's Disease drug candidate.
Mol Neurodegener, 18, 2023
|
|
8CDY
 
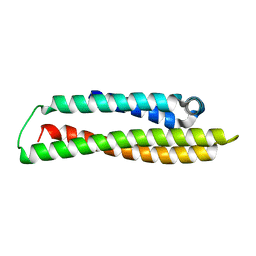 | | N-terminal domain of human apolipoprotein E | | Descriptor: | Maltodextrin-binding protein,Apolipoprotein E | | Authors: | Marek, M, Nemergut, M. | | Deposit date: | 2023-02-01 | | Release date: | 2023-07-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Domino-like effect of C112R mutation on ApoE4 aggregation and its reduction by Alzheimer's Disease drug candidate.
Mol Neurodegener, 18, 2023
|
|
8CKP
 
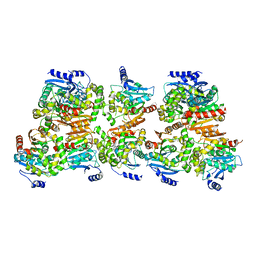 | | X-ray structure of the crystallization-prone form of subfamily III haloalkane dehalogenase DhmeA from Haloferax mediterranei | | Descriptor: | Alpha/beta fold hydrolase, CHLORIDE ION | | Authors: | Marek, M, Chmelova, K, Schenkmayerova, A, Croll, T, Read, R.J, Diederichs, K. | | Deposit date: | 2023-02-16 | | Release date: | 2023-08-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Multimeric structure of a subfamily III haloalkane dehalogenase-like enzyme solved by combination of cryo-EM and x-ray crystallography.
Protein Sci., 32, 2023
|
|
7Q1B
 
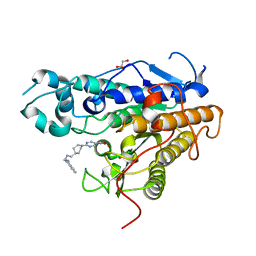 | | Crystal structure of Trypanosoma cruzi histone deacetylase DAC2 complexed with Quisinostat | | Descriptor: | 2-[4-[[(1-methylindol-3-yl)methylamino]methyl]piperidin-1-yl]-~{N}-oxidanyl-pyrimidine-5-carboxamide, GLYCEROL, Histone deacetylase DAC2, ... | | Authors: | Marek, M, Ramos-Morales, E, Romier, C. | | Deposit date: | 2021-10-18 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Species-selective targeting of pathogens revealed by the atypical structure and active site of Trypanosoma cruzi histone deacetylase DAC2.
Cell Rep, 37, 2021
|
|
6FU1
 
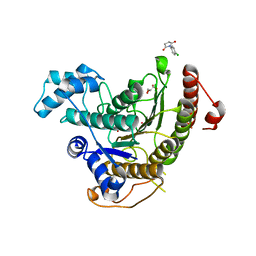 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a n-alkyl hydroxamate | | Descriptor: | GLYCEROL, Histone deacetylase, POTASSIUM ION, ... | | Authors: | Marek, M, Shaik, T.B, Romier, C. | | Deposit date: | 2018-02-26 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.548 Å) | | Cite: | A Novel Class of Schistosoma mansoni Histone Deacetylase 8 (HDAC8) Inhibitors Identified by Structure-Based Virtual Screening and In Vitro Testing.
Molecules, 23, 2018
|
|
4BZ8
 
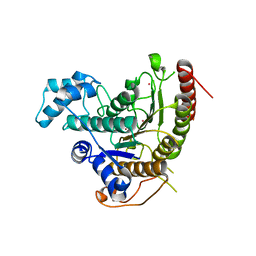 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with J1038 | | Descriptor: | (2R)-2-methyl-3-oxo-4H-1,4-benzothiazine-6-carbohydroxamic acid, HISTONE DEACETYLASE 8, POTASSIUM ION, ... | | Authors: | Marek, M, Romier, C. | | Deposit date: | 2013-07-24 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural Basis for the Inhibition of Histone Deacetylase 8 (Hdac8), a Key Epigenetic Player in the Blood Fluke Schistosoma Mansoni.
Plos Pathog., 9, 2013
|
|
4BZ7
 
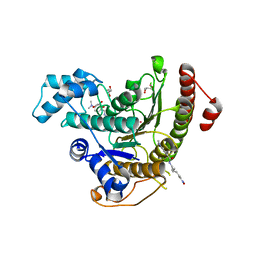 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with M344 | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, GLYCEROL, HISTONE DEACETYLASE 8, ... | | Authors: | Marek, M, Romier, C. | | Deposit date: | 2013-07-24 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for the Inhibition of Histone Deacetylase 8 (Hdac8), a Key Epigenetic Player in the Blood Fluke Schistosoma Mansoni.
Plos Pathog., 9, 2013
|
|
4BZ6
 
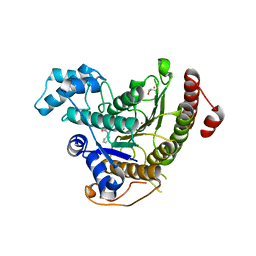 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with SAHA | | Descriptor: | DIMETHYLFORMAMIDE, GLYCEROL, HISTONE DEACETYLASE 8, ... | | Authors: | Marek, M, Romier, C. | | Deposit date: | 2013-07-24 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Inhibition of Histone Deacetylase 8 (Hdac8), a Key Epigenetic Player in the Blood Fluke Schistosoma Mansoni.
Plos Pathog., 9, 2013
|
|
4BZ9
 
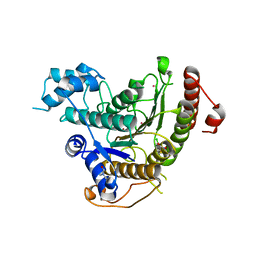 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with J1075 | | Descriptor: | 3-chlorobenzothiophene-2-carbohydroxamic acid, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Marek, M, Romier, C. | | Deposit date: | 2013-07-24 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Inhibition of Histone Deacetylase 8 (Hdac8), a Key Epigenetic Player in the Blood Fluke Schistosoma Mansoni.
Plos Pathog., 9, 2013
|
|
4BZ5
 
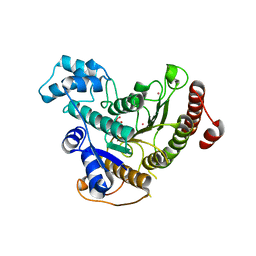 | | Crystal structure of Schistosoma mansoni HDAC8 | | Descriptor: | HISTONE DEACETYLASE 8, L(+)-TARTARIC ACID, POTASSIUM ION, ... | | Authors: | Marek, M, Romier, C. | | Deposit date: | 2013-07-24 | | Release date: | 2013-08-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.785 Å) | | Cite: | Structural Basis for the Inhibition of Histone Deacetylase 8 (Hdac8), a Key Epigenetic Player in the Blood Fluke Schistosoma Mansoni.
Plos Pathog., 9, 2013
|
|
4CQF
 
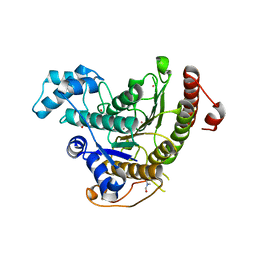 | |
6HQY
 
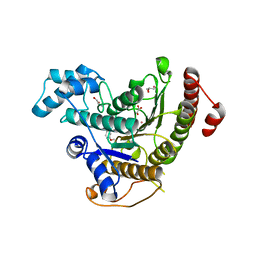 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with PCI-34051 | | Descriptor: | 1-[(4-methoxyphenyl)methyl]-~{N}-oxidanyl-indole-6-carboxamide, GLYCEROL, Histone deacetylase, ... | | Authors: | Marek, M, Shaik, T.B, Romier, C. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HSK
 
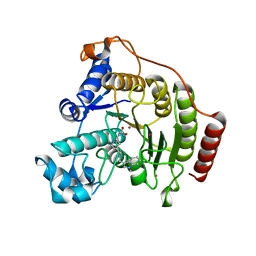 | | Crystal structure of a human HDAC8 L6 loop mutant complexed with Quisinostat | | Descriptor: | 2-[4-[[(1-methylindol-3-yl)methylamino]methyl]piperidin-1-yl]-~{N}-oxidanyl-pyrimidine-5-carboxamide, Histone deacetylase 8, POTASSIUM ION, ... | | Authors: | Marek, M, Shaik, T.B, Ramos-Morales, E, Romier, C. | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.096 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HSZ
 
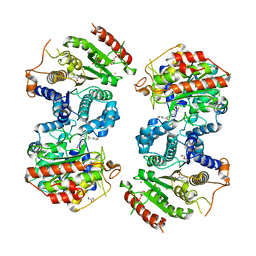 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 2 | | Descriptor: | 3-benzamido-4-methyl-~{N}-oxidanyl-benzamide, GLYCEROL, Histone deacetylase, ... | | Authors: | Marek, M, Shaik, T.B, Romier, C. | | Deposit date: | 2018-10-02 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.374 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HT8
 
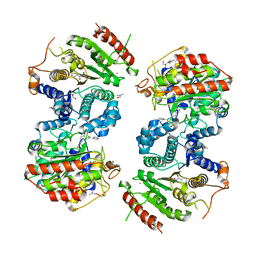 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 3 | | Descriptor: | 3-benzamido-4-methoxy-~{N}-oxidanyl-benzamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Marek, M, Shaik, T.B, Romier, C. | | Deposit date: | 2018-10-03 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.497 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6TY7
 
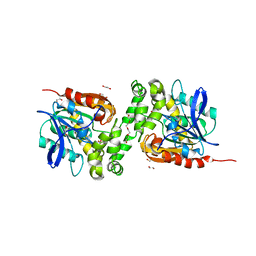 | | Crystal structure of haloalkane dehalogenase variant DhaA115 domain-swapped dimer type-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Markova, K, Chaloupkova, R, Damborsky, J, Marek, M. | | Deposit date: | 2020-01-15 | | Release date: | 2021-01-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Computational Enzyme Stabilization Can Affect Folding Energy Landscapes and Lead to Catalytically Enhanced Domain-Swapped Dimers
Acs Catalysis, 11, 2021
|
|
8OOH
 
 | |
