8QB5
 
 | | Crystal structure of apo-GltTk obtained with in meso crystallization (P6322 space group) | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, OLEIC ACID, Proton/glutamate symporter, ... | | Authors: | Marin, E, Guskov, A, Borshchevskiy, V, Kovalev, K. | | Deposit date: | 2023-08-24 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of apo-GltTk obtained with in meso crystallization (P6322 space group)
To Be Published
|
|
8QB4
 
 | |
9GBK
 
 | | Blm10-20S proteasome complex from pre1-1 | | Descriptor: | Probable proteasome subunit alpha type-7, Proteasome activator BLM10, Proteasome subunit alpha type-1, ... | | Authors: | Mark, E, Ramos, P.C, Kayser, F, Hoeckendorff, J, Dohmen, R.J, Wendler, P. | | Deposit date: | 2024-07-31 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Structural roles of Ump1 and beta-subunit propeptides in proteasome biogenesis.
Life Sci Alliance, 7, 2024
|
|
6MII
 
 | | Crystal structure of minichromosome maintenance protein MCM/DNA complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), MAGNESIUM ION, ... | | Authors: | Enemark, E.J, Meagher, M, Epling, L.B. | | Deposit date: | 2018-09-19 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | DNA translocation mechanism of the MCM complex and implications for replication initiation.
Nat Commun, 10, 2019
|
|
9MOJ
 
 | | Saccharolobus solfataricus GINS tetramer | | Descriptor: | GINS subunit domain-containing protein, SsoGINS51 | | Authors: | Enemark, E.J, Shankar, S. | | Deposit date: | 2024-12-26 | | Release date: | 2025-04-23 | | Last modified: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Saccharolobus solfataricus GINS tetramer.
Acta Crystallogr.,Sect.F, 81, 2025
|
|
7VUR
 
 | | Crystal structure of AlleyCat9 with calcium but no inhibitor | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, AlleyCat, CALCIUM ION | | Authors: | Margheritis, E, Takahashi, K, Korendovych, I.V, Tame, J.R.H. | | Deposit date: | 2021-11-04 | | Release date: | 2022-07-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | NMR-guided directed evolution.
Nature, 610, 2022
|
|
8RVO
 
 | | Proteasomal late precursor complex from pre1-1, state 1 | | Descriptor: | Probable proteasome subunit alpha type-7, Proteasome assembly chaperone 2, Proteasome chaperone 1, ... | | Authors: | Mark, E, Ramos, P.C, Kayser, F, Hoeckendorff, J, Dohmen, R.J, Wendler, P. | | Deposit date: | 2024-02-01 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structural roles of Ump1 and beta-subunit propeptides in proteasome biogenesis.
Life Sci Alliance, 7, 2024
|
|
8RVQ
 
 | | 20S proteasome from pre1-1 | | Descriptor: | Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, ... | | Authors: | Mark, E, Ramos, P.C, Kayser, F, Hoeckendorff, J, Dohmen, R.J, Wendler, P. | | Deposit date: | 2024-02-01 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.02 Å) | | Cite: | Structural roles of Ump1 and beta-subunit propeptides in proteasome biogenesis.
Life Sci Alliance, 7, 2024
|
|
8RVL
 
 | | Proteasomal late precursor complex from pre1-1 | | Descriptor: | Probable proteasome subunit alpha type-7, Proteasome assembly chaperone 2, Proteasome chaperone 1, ... | | Authors: | Mark, E, Ramos, P.C, Kayser, F, Hoeckendorff, J, Dohmen, R.J, Wendler, P. | | Deposit date: | 2024-02-01 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Structural roles of Ump1 and beta-subunit propeptides in proteasome biogenesis.
Life Sci Alliance, 7, 2024
|
|
8BBU
 
 | | Crystal structure of medical leech destabilase (high salt) | | Descriptor: | GLYCEROL, Lysozyme, MALONATE ION, ... | | Authors: | Marin, E, Bukhdruker, S, Manuvera, V, Kornilov, D, Zinovev, E, Bobrovsky, P, Lazarev, V, Borshchevskiy, V. | | Deposit date: | 2022-10-14 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural insights into thrombolytic activity of destabilase from medicinal leech.
Sci Rep, 13, 2023
|
|
8BBW
 
 | | Crystal structure of medical leech destabilase (low salt) | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme | | Authors: | Marin, E, Bukhdruker, S, Manuvera, V, Kornilov, D, Zinovev, E, Bobrovsky, P, Lazarev, V, Borshchevskiy, V. | | Deposit date: | 2022-10-14 | | Release date: | 2023-02-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural insights into thrombolytic activity of destabilase from medicinal leech.
Sci Rep, 13, 2023
|
|
8RVP
 
 | | Proteasomal late precursor complex from pre1-1, state 2 | | Descriptor: | Probable proteasome subunit alpha type-7, Proteasome assembly chaperone 2, Proteasome chaperone 1, ... | | Authors: | Mark, E, Ramos, P.C, Kayser, F, Hoeckendorff, J, Dohmen, R.J, Wendler, P. | | Deposit date: | 2024-02-01 | | Release date: | 2024-09-11 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.28 Å) | | Cite: | Structural roles of Ump1 and beta-subunit propeptides in proteasome biogenesis.
Life Sci Alliance, 7, 2024
|
|
7LIU
 
 | | DDX3X bound to ATP analog and remodeled RNA:DNA hybrid | | Descriptor: | 5'-R(*GP*GP*GP*CP*GP*GP*G)-D(P*CP*CP*CP*GP*CP*CP*C)-3', ATP-dependent RNA helicase DDX3X, MAGNESIUM ION, ... | | Authors: | Enemark, E.J, Yu, S. | | Deposit date: | 2021-01-27 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | DDX3X bound to ATP analog and remodeled RNA:DNA hybrid
To Be Published
|
|
4YGI
 
 | |
6RQR
 
 | | Extended NHERF1 PDZ2 domain in complex with the PDZ-binding motif of CFTR | | Descriptor: | Na(+)/H(+) exchange regulatory cofactor NHE-RF1,Cystic fibrosis transmembrane conductance regulator | | Authors: | Martin, E.R, Ford, R.C, Robinson, R.C. | | Deposit date: | 2019-05-16 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | In vivocrystals reveal critical features of the interaction between cystic fibrosis transmembrane conductance regulator (CFTR) and the PDZ2 domain of Na+/H+exchange cofactor NHERF1.
J.Biol.Chem., 295, 2020
|
|
1F08
 
 | | CRYSTAL STRUCTURE OF THE DNA-BINDING DOMAIN OF THE REPLICATION INITIATION PROTEIN E1 FROM PAPILLOMAVIRUS | | Descriptor: | BROMIDE ION, REPLICATION PROTEIN E1 | | Authors: | Enemark, E.J, Chen, G, Vaughn, D.E, Stenlund, A, Joshua-Tor, L. | | Deposit date: | 2000-05-15 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the DNA binding domain of the replication initiation protein E1 from papillomavirus.
Mol.Cell, 6, 2000
|
|
6EE5
 
 | |
4EB5
 
 | | A. fulgidus IscS-IscU complex structure | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, ... | | Authors: | Marinoni, E.N, de Oliveira, J.S, Nicolet, Y, Raulfs, E.C, Amara, P, Dean, D.R, Fontecilla-Camps, J.C. | | Deposit date: | 2012-03-23 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | (IscS-IscU)2 complex structures provide insights into Fe2S2 biogenesis and transfer.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
1CHK
 
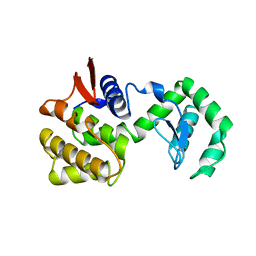 | |
6E5C
 
 | | Solution NMR structure of a de novo designed double-stranded beta-helix | | Descriptor: | De novo beta protein | | Authors: | Marcos, E, Chidyausiku, T.M, McShan, A, Evangelidis, T, Nerli, S, Sgourakis, N, Tripsianes, K, Baker, D. | | Deposit date: | 2018-07-19 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | De novo design of a non-local beta-sheet protein with high stability and accuracy.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4EB7
 
 | | A. fulgidus IscS-IscU complex structure | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FE2/S2 (INORGANIC) CLUSTER, GLYCEROL, ... | | Authors: | Marinoni, E.N, de Oliveira, J.S, Nicolet, Y, Raulfs, E.C, Amara, P, Dean, D.R, Fontecilla-Camps, J.C. | | Deposit date: | 2012-03-23 | | Release date: | 2012-05-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | (IscS-IscU)2 complex structures provide insights into Fe2S2 biogenesis and transfer.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
1KSY
 
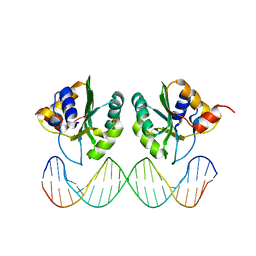 | | Crystal Structures of Two Intermediates in the Assembly of the Papillomavirus Replication Initiation Complex | | Descriptor: | E1 Recognition Sequence, Strand 1, Strand 2, ... | | Authors: | Enemark, E.J, Stenlund, A, Joshua-Tor, L. | | Deposit date: | 2002-01-14 | | Release date: | 2002-03-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structures of two intermediates in the assembly of the papillomavirus replication initiation complex.
EMBO J., 21, 2002
|
|
2Z79
 
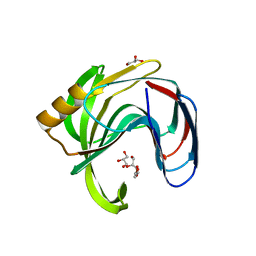 | | High resolution crystal structure of a glycoside hydrolase family 11 xylanase of Bacillus subtilis | | Descriptor: | Endo-1,4-beta-xylanase A, GLYCEROL | | Authors: | Vandermarliere, E, Bourgois, T.M, Strelkov, S.V, Delcour, J.A, Courtin, C.M, Rabijns, A. | | Deposit date: | 2007-08-16 | | Release date: | 2007-12-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystallographic analysis shows substrate binding at the -3 to +1 active-site subsites and at the surface of glycoside hydrolase family 11 endo-1,4-beta-xylanases.
Biochem.J., 410, 2008
|
|
1KSX
 
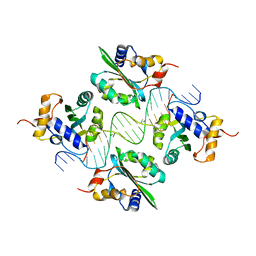 | |
1HN3
 
 | | SOLUTION STRUCTURE OF THE N-TERMINAL 37 AMINO ACIDS OF THE MOUSE ARF TUMOR SUPPRESSOR PROTEIN | | Descriptor: | P19 ARF PROTEIN | | Authors: | DiGiammarino, E.L, Filippov, I, Weber, J.D, Bothner, B, Kriwacki, R.W. | | Deposit date: | 2000-12-05 | | Release date: | 2001-12-05 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the p53 regulatory domain of the p19Arf tumor suppressor protein.
Biochemistry, 40, 2001
|
|
