4WWB
 
 | | High-resolution structure of the Ni-bound form of the Y135F mutant of C. metallidurans CnrXs | | Descriptor: | CARBON DIOXIDE, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Volbeda, A, Coves, J, Maillard, A.P, Kuennemann, S, Grosse, C, Schleuder, G, Petit-Haertlein, I, de Rosny, E, Nies, D.H. | | Deposit date: | 2014-11-10 | | Release date: | 2015-02-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | Response of CnrX from Cupriavidus metallidurans CH34 to nickel binding.
Metallomics, 7, 2015
|
|
2YJP
 
 | | Crystal structure of the solute receptors for L-cysteine of Neisseria gonorrhoeae | | Descriptor: | 1,2-ETHANEDIOL, CYSTEINE, PUTATIVE ABC TRANSPORTER, ... | | Authors: | Bulut, H, Moniot, S, Scheffel, F, Gathmann, S, Licht, A, Saenger, W, Schneider, E. | | Deposit date: | 2011-05-23 | | Release date: | 2011-12-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal Structures of Two Solute Receptors for L-Cystine and L-Cysteine, Respectively, of the Human Pathogen Neisseria Gonorrhoeae.
J.Mol.Biol., 415, 2012
|
|
6OY4
 
 | | Crystal structure of complex between recombinant Der p 2.0103 and Fab fragment of 7A1 | | Descriptor: | Der p 2 variant 3, Fab fragment of IgG, HEAVY CHAIN, ... | | Authors: | Kapingidza, A.B, Offermann, L.R, Glesner, J, Wunschmann, S, Vailes, L.D, Chapman, M.D.C, Pomes, A, Chruszcz, M. | | Deposit date: | 2019-05-14 | | Release date: | 2019-08-28 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Human IgE Antibody Binding Site on Der p 2 for the Design of a Recombinant Allergen for Immunotherapy.
J Immunol., 203, 2019
|
|
3IZJ
 
 | | Mm-cpn rls with ATP and AlFx | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
1PJA
 
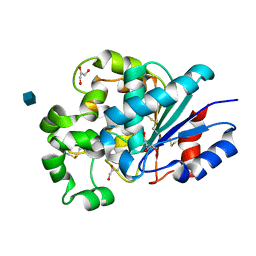 | | The crystal structure of palmitoyl protein thioesterase-2 reveals the basis for divergent substrate specificities of the two lysosomal thioesterases (PPT1 and PPT2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Palmitoyl-protein thioesterase 2 precursor | | Authors: | Calero, G, Gupta, P, Nonato, M.C, Tandel, S, Biehl, E.R, Hofmann, S.L, Clardy, J. | | Deposit date: | 2003-06-02 | | Release date: | 2003-09-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of palmitoyl protein thioesterase-2 (PPT2) reveals the basis for divergent substrate specificities of the two lysosomal thioesterases, PPT1 and PPT2.
J.Biol.Chem., 278, 2003
|
|
3IZI
 
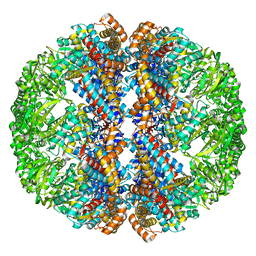 | | Mm-cpn rls with ATP | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3IZH
 
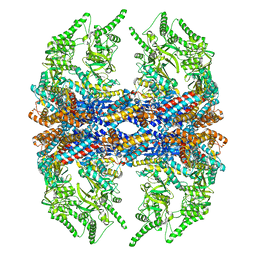 | | Mm-cpn D386A with ATP | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3IZM
 
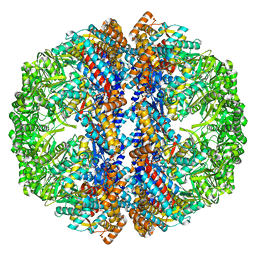 | | Mm-cpn wildtype with ATP | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-30 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (7.2 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3IZL
 
 | | Mm-cpn rls deltalid with ATP and AlFx | | Descriptor: | Mm-cpn rls deltalid | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3IZN
 
 | | Mm-cpn deltalid with ATP | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-30 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
3IZK
 
 | | Mm-cpn rls deltalid with ATP | | Descriptor: | Chaperonin | | Authors: | Douglas, N.R, Reissmann, S, Zhang, J, Chen, B, Jakana, J, Kumar, R, Chiu, W, Frydman, J. | | Deposit date: | 2010-10-29 | | Release date: | 2011-02-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Dual Action of ATP Hydrolysis Couples Lid Closure to Substrate Release into the Group II Chaperonin Chamber.
Cell(Cambridge,Mass.), 144, 2011
|
|
1PVW
 
 | | 3,4-dihydroxy-2-butanone 4-phosphate synthase from M. jannaschii | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, CALCIUM ION, PHOSPHATE ION, ... | | Authors: | Steinbacher, S, Schiffmann, S, Richter, G, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2003-06-29 | | Release date: | 2003-11-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of 3,4-Dihydroxy-2-butanone 4-Phosphate Synthase from Methanococcus jannaschii in
Complex with Divalent Metal Ions and the Substrate Ribulose 5-Phosphate: IMPLICATIONS FOR THE
CATALYTIC MECHANISM
J.Biol.Chem., 278, 2003
|
|
1PVY
 
 | | 3,4-dihydroxy-2-butanone 4-phosphate synthase from M. jannaschii in complex with ribulose 5-phosphate | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, CALCIUM ION, RIBULOSE-5-PHOSPHATE, ... | | Authors: | Steinbacher, S, Schiffmann, S, Richter, G, Huber, R, Bacher, A, Fischer, M. | | Deposit date: | 2003-06-29 | | Release date: | 2003-11-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of 3,4-Dihydroxy-2-butanone 4-Phosphate Synthase from Methanococcus jannaschii in
Complex with Divalent Metal Ions and the Substrate Ribulose 5-Phosphate: IMPLICATIONS FOR THE
CATALYTIC MECHANISM
J.Biol.Chem., 278, 2003
|
|
1AG2
 
 | | PRION PROTEIN DOMAIN PRP(121-231) FROM MOUSE, NMR, 2 MINIMIZED AVERAGE STRUCTURE | | Descriptor: | MAJOR PRION PROTEIN | | Authors: | Billeter, M, Riek, R, Wider, G, Wuthrich, K, Hornemann, S, Glockshuber, R. | | Deposit date: | 1997-03-31 | | Release date: | 1997-10-08 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the mouse prion protein domain PrP(121-231).
Nature, 382, 1996
|
|
2YEN
 
 | | Solution structure of the skeletal muscle and neuronal voltage gated sodium channel antagonist mu-conotoxin CnIIIC | | Descriptor: | Mu-conotoxin CnIIIC | | Authors: | Favreau, P, Benoit, E, Hocking, H.G, Carlier, L, D'hoedt, D, Leipold, E, Markgraf, R, Schlumberger, S, Cordova, M.A, Gaertner, H, Paolini-Bertrand, M, Hartley, O, Tytgat, J, Heinemann, S.H, Bertrand, D, Boelens, R, Stocklin, R, Molgo, J. | | Deposit date: | 2011-03-28 | | Release date: | 2012-02-08 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Novel Mu-Conopeptide, Cniiic, Exerts Potent and Preferential Inhibition of Na(V) 1.2/1.4 Channels and Blocks Neuronal Nicotinic Acetylcholine Receptors.
Br.J.Pharmacol., 166, 2012
|
|
1YG9
 
 | | The structure of mutant (N93Q) of bla g 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Aspartic protease Bla g 2, ... | | Authors: | Li, M, Gustchina, A, Wuenschmann, S, Pomes, A, Wlodawer, A. | | Deposit date: | 2005-01-04 | | Release date: | 2005-03-22 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of cockroach allergen Bla g 2, an unusual zinc binding aspartic protease with a novel mode of self-inhibition.
J.Mol.Biol., 348, 2005
|
|
3O8W
 
 | | Archaeoglobus fulgidus GlnK1 | | Descriptor: | ACETATE ION, Nitrogen regulatory protein P-II (GlnB-1) | | Authors: | Litz, C, Helfmann, S, Gerhardt, S, Andrade, S.L.A. | | Deposit date: | 2010-08-03 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure of GlnK1, a signalling protein from Archaeoglobus fulgidus.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
4ZPQ
 
 | | Crystal Structure of Protocadherin Gamma C5 EC1-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MCG133388, ... | | Authors: | Wolcott, H.N, Goodman, K.M, Bahna, F, Mannepalli, S, Shapiro, L. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.099 Å) | | Cite: | Molecular Logic of Neuronal Self-Recognition through Protocadherin Domain Interactions.
Cell, 163, 2015
|
|
4ZPM
 
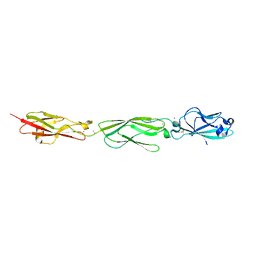 | | Crystal Structure of Protocadherin Alpha C2 EC1-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protein Pcdhac2, ... | | Authors: | Goodman, K.M, Mannepalli, S, Shapiro, L. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular Logic of Neuronal Self-Recognition through Protocadherin Domain Interactions.
Cell, 163, 2015
|
|
6XU1
 
 | | Crystal structure of tetrameric human H215A-SAMHD1 (residues 109-626) with GTP, dAMPNPP and Mg | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, ... | | Authors: | Morris, E.R, Kunzelmann, S, Caswell, S.J, Arnold, L.H, Purkiss, A.G, Kelly, G, Taylor, I.A. | | Deposit date: | 2020-01-17 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of SAMHD1 inhibitor complexes reveal the mechanism of water-mediated dNTP hydrolysis.
Nat Commun, 11, 2020
|
|
3O3L
 
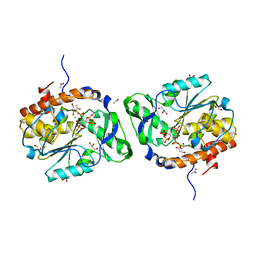 | | Structure of the PTP-like phytase from Selenomonas ruminantium in complex with myo-inositol (1,3,4,5)tetrakisphosphate | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Gruninger, R.J, Selinger, L.B, Mosimann, S.C. | | Deposit date: | 2010-07-25 | | Release date: | 2011-12-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural analysis of substrate binding in PTPLPs
To be Published
|
|
6YOM
 
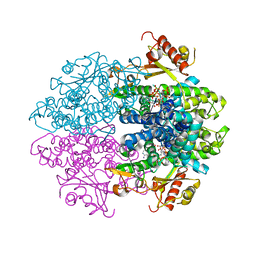 | | Crystal structure of tetrameric human D137N-SAMHD1 (residues 109-626) with XTP, dATP, dCMPNPP, Mn and Mg | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, ... | | Authors: | Morris, E.R, Kunzelmann, S, Caswell, S.J, Arnold, L.H, Purkiss, A.G, Kelly, G, Taylor, I.A. | | Deposit date: | 2020-04-14 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structures of SAMHD1 inhibitor complexes reveal the mechanism of water-mediated dNTP hydrolysis.
Nat Commun, 11, 2020
|
|
3M0E
 
 | | Crystal structure of the ATP-bound state of Walker B mutant of NtrC1 ATPase domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Transcriptional regulator (NtrC family) | | Authors: | Chen, B, Sysoeva, T.A, Chowdhury, S, Rusu, M, Birmanns, S, Guo, L, Hanson, J, Yang, H, Nixon, B.T. | | Deposit date: | 2010-03-02 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Engagement of Arginine Finger to ATP Triggers Large Conformational Changes in NtrC1 AAA+ ATPase for Remodeling Bacterial RNA Polymerase.
Structure, 18, 2010
|
|
1W6T
 
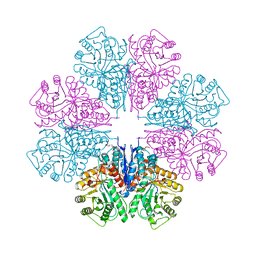 | | Crystal Structure Of Octameric Enolase From Streptococcus pneumoniae | | Descriptor: | ENOLASE, MAGNESIUM ION, NONAETHYLENE GLYCOL | | Authors: | Ehinger, S, Schubert, W.-D, Bergmann, S, Hammerschmidt, S, Heinz, D.W. | | Deposit date: | 2004-08-24 | | Release date: | 2005-08-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Plasmin(Ogen)-Binding Alpha-Enolase from Streptococcus Pneumoniae: Crystal Structure and Evaluation of Plasmin(Ogen)-Binding Sites
J.Mol.Biol., 343, 2004
|
|
4ZPP
 
 | | Crystal Structure of Protocadherin Gamma C5 EC1-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MCG133388, ... | | Authors: | Wolcott, H.N, Goodman, K.M, Bahna, F, Mannepalli, S, Shapiro, L. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Molecular Logic of Neuronal Self-Recognition through Protocadherin Domain Interactions.
Cell, 163, 2015
|
|
