7A5Y
 
 | | Crystal structure of tetrameric human H215A-SAMHD1 (residues 109-626) with Rp-dGTP-alphaS (T8T) and Mg | | Descriptor: | 2'-deoxyguanosine-5'-O-(1-thiotriphosphate), Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, FE (III) ION, ... | | Authors: | Morris, E.R, Kunzelmann, S, Caswell, S.J, Purkiss, A, Taylor, I.A. | | Deposit date: | 2020-08-24 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Probing the Catalytic Mechanism and Inhibition of SAMHD1 Using the Differential Properties of R p - and S p -dNTP alpha S Diastereomers.
Biochemistry, 60, 2021
|
|
1W3Q
 
 | | NimA from D. radiodurans with covalenly bound lactate | | Descriptor: | ACETATE ION, LACTIC ACID, NIMA-RELATED PROTEIN | | Authors: | Leiros, H.-K.S, Kozielski-Stuhrmann, S, Kapp, U, Terradot, L, Leonard, G.A, Mcsweeney, S.M. | | Deposit date: | 2004-07-17 | | Release date: | 2004-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural Basis of 5-Nitroimidazole Antibiotic Resistance: The Crystal Structure of Nima from Deinococcus Radiodurans
J.Biol.Chem., 279, 2004
|
|
1W3O
 
 | | Crystal structure of NimA from D. radiodurans | | Descriptor: | ACETATE ION, NIMA-RELATED PROTEIN, PYRUVIC ACID | | Authors: | Leiros, H.-K.S, Kozielski-Stuhrmann, S, Kapp, U, Terradot, L, Leonard, G.A, McSweeney, S.M. | | Deposit date: | 2004-07-17 | | Release date: | 2004-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of 5-Nitroimidazole Antibiotic Resistance: The Crystal Structure of Nima from Deinococcus Radiodurans
J.Biol.Chem., 279, 2004
|
|
1W3P
 
 | | NimA from D. radiodurans with a His71-Pyruvate residue | | Descriptor: | ACETATE ION, NIMA-RELATED PROTEIN, PYRUVIC ACID | | Authors: | Leiros, H.-K.S, Kozielski-Stuhrmann, S, Kapp, U, Terradot, L, Leonard, G.A, McSweeney, S.M. | | Deposit date: | 2004-07-17 | | Release date: | 2004-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of 5-Nitroimidazole Antibiotic Resistance: The Crystal Structure of Nima from Deinococcus Radiodurans
J.Biol.Chem., 279, 2004
|
|
1W3R
 
 | | NimA from D. radiodurans with Metronidazole and Pyruvate | | Descriptor: | ACETATE ION, Metronidazole, NIMA-RELATED PROTEIN, ... | | Authors: | Leiros, H.-K.S, Kozielski-Stuhrmann, S, Kapp, U, Terradot, L, Leonard, G.A, McSweeney, S.M. | | Deposit date: | 2004-07-17 | | Release date: | 2004-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis of 5-Nitroimidazole Antibiotic Resistance: The Crystal Structure of Nima from Deinococcus Radiodurans
J.Biol.Chem., 279, 2004
|
|
6ZTB
 
 | | Crystal Structure of human P-Cadherin EC1_EC2 | | Descriptor: | CALCIUM ION, Cadherin-3, SODIUM ION | | Authors: | Rondeau, J.M, Lehmann, S. | | Deposit date: | 2020-07-17 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | PCA062, a P-cadherin Targeting Antibody-Drug Conjugate, Displays Potent Antitumor Activity Against P-cadherin-expressing Malignancies.
Mol.Cancer Ther., 20, 2021
|
|
6ZTR
 
 | | Crystal Structure of the anti-human P-Cadherin Fab CQY684 in complex with human P-Cadherin(108-324) | | Descriptor: | CALCIUM ION, CQY684 Fab heavy-chain, CQY684 Fab light-chain, ... | | Authors: | Rondeau, J.M, Lehmann, S. | | Deposit date: | 2020-07-20 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | PCA062, a P-cadherin Targeting Antibody-Drug Conjugate, Displays Potent Antitumor Activity Against P-cadherin-expressing Malignancies.
Mol.Cancer Ther., 20, 2021
|
|
6ZTF
 
 | | Crystal Structure of the anti-human P-Cadherin Fab CQY684 | | Descriptor: | CQY684 Fab heavy-chain, CQY684 Fab light-chain | | Authors: | Rondeau, J.M, Lehmann, S. | | Deposit date: | 2020-07-20 | | Release date: | 2021-05-05 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | PCA062, a P-cadherin Targeting Antibody-Drug Conjugate, Displays Potent Antitumor Activity Against P-cadherin-expressing Malignancies.
Mol.Cancer Ther., 20, 2021
|
|
6ZWT
 
 | | Crystal structure of DNA-binding domain of OmpR of two-component system of Acinetobacter baumannii | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, SULFATE ION, ... | | Authors: | Narwal, M, Lucchini, V, Trebosc, V, Gartenmann, S, Pieren, M, Kemmer, C, Kammerer, R.A. | | Deposit date: | 2020-07-28 | | Release date: | 2021-08-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Targeting virulence regulation to disarm Acinetobacter baumannii pathogenesis.
Virulence, 13, 2022
|
|
7AO7
 
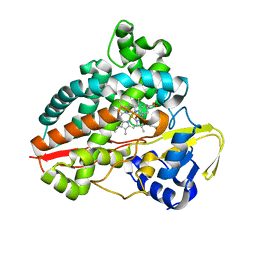 | | Structure of CYP153A from Polaromonas sp. in complex with octan-1-ol | | Descriptor: | Cytochrome P450, OCTAN-1-OL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Zukic, E, Rowlinson, B, Sharma, M, Hoffmann, S, Hauer, B, Grogan, G. | | Deposit date: | 2020-10-13 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Substrate Anchoring and Flexibility Reduction in CYP153AM.aq Leads to Highly Improved Efficiency toward Octanoic Acid
Acs Catalysis, 11, 2021
|
|
4WTY
 
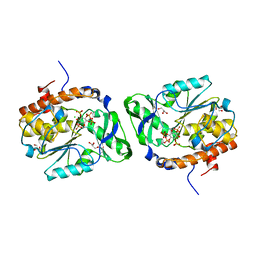 | | Structure of the PTP-like myo-inositol phosphatase from Selenomonas ruminantium in complex with myo-inositol-(1,3,4,5)-tetrakisphosphate | | Descriptor: | CHLORIDE ION, GLYCEROL, INOSITOL-(1,3,4,5)-TETRAKISPHOSPHATE, ... | | Authors: | Bruder, L.M, Mosimann, S.C. | | Deposit date: | 2014-10-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the PTP-like phytase from Selenomonas ruminantium in complex with myo-inositol-(1,3,4,5)-tetrakisphosphate
To Be Published
|
|
1JG5
 
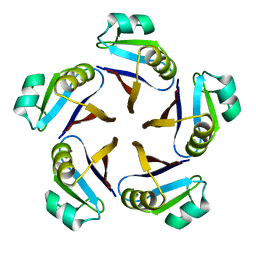 | | CRYSTAL STRUCTURE OF RAT GTP CYCLOHYDROLASE I FEEDBACK REGULATORY PROTEIN, GFRP | | Descriptor: | GTP CYCLOHYDROLASE I FEEDBACK REGULATORY PROTEIN, POTASSIUM ION | | Authors: | Bader, G, Schiffmann, S, Herrmann, A, Fischer, M, Gutlich, M, Auerbach, G, Ploom, T, Bacher, A, Huber, R, Lemm, T. | | Deposit date: | 2001-06-23 | | Release date: | 2001-10-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of rat GTP cyclohydrolase I feedback regulatory protein, GFRP.
J.Mol.Biol., 312, 2001
|
|
4P86
 
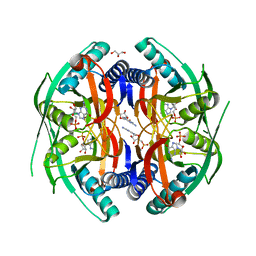 | | Structure of PyrR protein from Bacillus subtilis with GMP | | Descriptor: | Bifunctional protein PyrR, GLYCEROL, GUANOSINE-5'-MONOPHOSPHATE | | Authors: | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | Deposit date: | 2014-03-30 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
4P84
 
 | | Structure of engineered PyrR protein (VIOLET PyrR) | | Descriptor: | Bifunctional protein PyrR, GLYCEROL, SULFATE ION | | Authors: | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | Deposit date: | 2014-03-30 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
4P80
 
 | | Structure of ancestral PyrR protein (AncGREENPyrR) | | Descriptor: | Ancestral PyrR protein (Green), SULFATE ION | | Authors: | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | Deposit date: | 2014-03-29 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
2MUS
 
 | | HADDOCK calculated model of LIN5001 bound to the HET-s amyloid | | Descriptor: | 3''',4'-bis(carboxymethyl)-2,2':5',2'':5'',2''':5''',2''''-quinquethiophene-5,5''''-dicarboxylic acid, Heterokaryon incompatibility protein s | | Authors: | Hermann, U.S, Schuetz, A.K, Shirani, H, Saban, D, Nuvolone, M, Huang, D.H, Li, B, Ballmer, B, Aslund, A.K.O, Mason, J.J, Rushing, E, Budka, H, Hammarstrom, P, Bockmann, A, Caflisch, A, Meier, B.H, Nilsson, P.K.R, Hornemann, S, Aguzzi, A. | | Deposit date: | 2014-09-16 | | Release date: | 2017-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-based drug design identifies polythiophenes as antiprion compounds.
Sci Transl Med, 7, 2015
|
|
6SKY
 
 | | FAT and kinase domain of CtTel1 | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase Tel1 | | Authors: | Jansma, M, Eustermann, S.E, Kostrewa, D, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-08-16 | | Release date: | 2019-10-30 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Near-Complete Structure and Model of Tel1ATM from Chaetomium thermophilum Reveals a Robust Autoinhibited ATP State.
Structure, 28, 2020
|
|
6SL0
 
 | | Complete CtTel1 dimer with C2 symmetry | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase Tel1 | | Authors: | Jansma, M, Eustermann, S.E, Kostrewa, D, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-08-16 | | Release date: | 2019-10-30 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Near-Complete Structure and Model of Tel1ATM from Chaetomium thermophilum Reveals a Robust Autoinhibited ATP State.
Structure, 28, 2020
|
|
4P3K
 
 | | Structure of ancestral PyrR protein (PLUMPyrR) | | Descriptor: | Ancestral PyrR protein (Plum), PENTAETHYLENE GLYCOL, SODIUM ION, ... | | Authors: | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | Deposit date: | 2014-03-08 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
4WU2
 
 | | Structure of the PTP-like myo-inositol phosphatase from Selenomonas ruminantium in complex with myo-inositol-(1,4,5)-trikisphosphate | | Descriptor: | CHLORIDE ION, D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Bruder, L.M, Mosimann, S.C. | | Deposit date: | 2014-10-30 | | Release date: | 2015-11-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the PTP-like phytase from Selenomonas ruminantium in complex with myo-inositol-(1,4,5)-trikisphosphate
To Be Published
|
|
4P81
 
 | | Structure of ancestral PyrR protein (AncORANGEPyrR) | | Descriptor: | Ancestral PyrR protein (Orange), GLYCEROL, SULFATE ION | | Authors: | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | Deposit date: | 2014-03-29 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
4P82
 
 | | Structure of PyrR protein from Bacillus subtilis | | Descriptor: | Bifunctional protein PyrR, SULFATE ION | | Authors: | Perica, T, Kondo, Y, Tiwari, S, McLaughlin, S, Steward, A, Reuter, N, Clarke, J, Teichmann, S.A. | | Deposit date: | 2014-03-30 | | Release date: | 2014-12-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Evolution of oligomeric state through allosteric pathways that mimic ligand binding.
Science, 346, 2014
|
|
6SL1
 
 | | Structure of the open conformation of CtTel1 | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase Tel1 | | Authors: | Jansma, M, Eustermann, S.E, Kostrewa, D, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-08-16 | | Release date: | 2019-10-30 | | Last modified: | 2020-05-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Near-Complete Structure and Model of Tel1ATM from Chaetomium thermophilum Reveals a Robust Autoinhibited ATP State.
Structure, 28, 2020
|
|
6SKZ
 
 | | Structure of the closed conformation of CtTel1 | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase Tel1 | | Authors: | Jansma, M, Eustermann, S.E, Kostrewa, D, Lammens, K, Hopfner, K.P. | | Deposit date: | 2019-08-16 | | Release date: | 2019-10-30 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Near-Complete Structure and Model of Tel1ATM from Chaetomium thermophilum Reveals a Robust Autoinhibited ATP State.
Structure, 28, 2020
|
|
6TFP
 
 | | BTK in complex with LOU064, a potent and highly selective covalent inhibitor | | Descriptor: | SODIUM ION, Tyrosine-protein kinase BTK, ~{N}-[3-[6-azanyl-5-[2-[methyl(propanoyl)amino]ethoxy]pyrimidin-4-yl]-5-fluoranyl-2-methyl-phenyl]-4-cyclopropyl-2-fluoranyl-benzamide | | Authors: | Scheufler, C, Hinniger, A, Gutmann, S. | | Deposit date: | 2019-11-14 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of LOU064 (Remibrutinib), a Potent and Highly Selective Covalent Inhibitor of Bruton's Tyrosine Kinase.
J.Med.Chem., 63, 2020
|
|
