6RXV
 
 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state B2 | | Descriptor: | 35S ribosomal RNA, 40S ribosomal protein S1, 40S ribosomal protein S11-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2019-10-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
3ELV
 
 | | Crystal Structure of Full-Length Yeast Pml1p | | Descriptor: | Pre-mRNA leakage protein 1, SULFATE ION | | Authors: | Trowitzsch, S, Weber, G, L hrmann, R, Wahl, M.C. | | Deposit date: | 2008-09-23 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the Pml1p subunit of the yeast precursor mRNA retention and splicing complex.
J.Mol.Biol., 385, 2009
|
|
6Q6F
 
 | | Crystal structure of IDH1 R132H in complex with HMS101 | | Descriptor: | (2~{R})-2-[2-[(3~{R})-3-(4-fluorophenyl)pyrrolidin-1-yl]ethyl]-1,4-dimethyl-piperazine, Isocitrate dehydrogenase [NADP] cytoplasmic | | Authors: | Chaturvedi, A, Goparaju, R, Gupta, C, Kluenemann, T, Araujo Cruz, M.M, Kloos, A, Goerlich, K, Schottmann, R, Struys, E.A, Ganser, A, Preller, M, Heuser, M. | | Deposit date: | 2018-12-10 | | Release date: | 2019-10-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | In vivo efficacy of mutant IDH1 inhibitor HMS-101 and structural resolution of distinct binding site.
Leukemia, 34, 2020
|
|
3QUK
 
 | | Crystal structures of the murine class I major histocompatibility complex H-2Db in complex with LCMV-derived gp33 altered peptide ligand (Y4A) | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Allerbring, E, Duru, A.D, Uchtenhagen, H, Madhurantakam, C, Grimm, S, Tomek, M.B, Mazumdar, P.A, Spetz, A, Friemann, R, Sandalova, T, Uhlin, M, Nygren, P, Achour, A. | | Deposit date: | 2011-02-24 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Unexpected T-cell recognition of an altered peptide ligand is driven by reversed thermodynamics.
Eur.J.Immunol., 42, 2012
|
|
3CKE
 
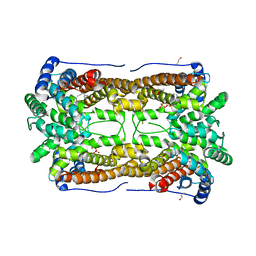 | | Crystal structure of aristolochene synthase in complex with 12,13-difluorofarnesyl diphosphate | | Descriptor: | (2E,6E)-12-fluoro-11-(fluoromethyl)-3,7-dimethyldodeca-2,6,10-trien-1-yl trihydrogen diphosphate, Aristolochene synthase, BETA-MERCAPTOETHANOL, ... | | Authors: | Shishova, E.Y, Yu, F, Miller, D.J, Faraldos, J.A, Zhao, Y, Coates, R.M, Allemann, R.K, Cane, D.E, Christianson, D.W. | | Deposit date: | 2008-03-14 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray Crystallographic Studies of Substrate Binding to Aristolochene Synthase Suggest a Metal Ion Binding Sequence for Catalysis.
J.Biol.Chem., 283, 2008
|
|
6SNT
 
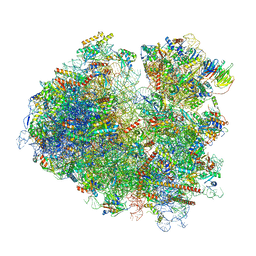 | | Yeast 80S ribosome stalled on SDD1 mRNA. | | Descriptor: | 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, 40S ribosomal protein S10-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Becker, T, Beckmann, R. | | Deposit date: | 2019-08-27 | | Release date: | 2020-03-04 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | RQT complex dissociates ribosomes collided on endogenous RQC substrate SDD1.
Nat.Struct.Mol.Biol., 27, 2020
|
|
2OG4
 
 | |
3QUL
 
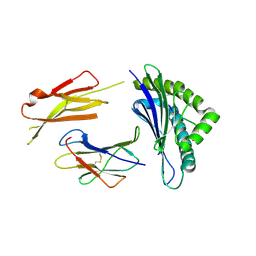 | | Crystal structures of the murine class I major histocompatibility complex H-2Db in complex with LCMV-derived gp33 altered peptide ligand (Y4S) | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Allerbring, E, Duru, A.D, Uchtenhagen, H, Madhurantakam, C, Grimm, S, Tomek, M.B, Mazumdar, P.A, Spetz, A, Friemann, R, Sandalova, T, Uhlin, M, Nygren, P, Achour, A. | | Deposit date: | 2011-02-24 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unexpected T-cell recognition of an altered peptide ligand is driven by reversed thermodynamics.
Eur.J.Immunol., 42, 2012
|
|
6SV4
 
 | | The cryo-EM structure of SDD1-stalled collided trisome. | | Descriptor: | 18S rRNA, 25S rRNA, 40S ribosomal protein S0-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Becker, T, Beckmann, R. | | Deposit date: | 2019-09-17 | | Release date: | 2020-03-04 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | RQT complex dissociates ribosomes collided on endogenous RQC substrate SDD1.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8C0V
 
 | | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate in single seam state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ruettermann, M, Koci, M, Lill, P, Geladas, E.D, Kaschani, F, Klink, B.U, Erdmann, R, Gatsogiannis, C. | | Deposit date: | 2022-12-19 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate.
Nat Commun, 14, 2023
|
|
8C0W
 
 | | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate in twin seam state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Ruettermann, M, Koci, M, Lill, P, Geladas, E.D, Kaschani, F, Klink, B.U, Erdmann, R, Gatsogiannis, C. | | Deposit date: | 2022-12-19 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure of the peroxisomal Pex1/Pex6 ATPase complex bound to a substrate.
Nat Commun, 14, 2023
|
|
6RXZ
 
 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state b | | Descriptor: | 35S ribosomal RNA, 40S ribosomal protein S11-like protein, 40S ribosomal protein S13-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
6RXT
 
 | | Cryo-EM structure of the 90S pre-ribosome (Kre33-Noc4) from Chaetomium thermophilum, state A | | Descriptor: | 35S ribosomal RNA, 40S ribosomal protein S1, 40S ribosomal protein S13-like protein, ... | | Authors: | Cheng, J, Kellner, N, Griesel, S, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2019-06-10 | | Release date: | 2019-08-14 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (7 Å) | | Cite: | Thermophile 90S Pre-ribosome Structures Reveal the Reverse Order of Co-transcriptional 18S rRNA Subdomain Integration.
Mol.Cell, 75, 2019
|
|
6TB3
 
 | | yeast 80S ribosome in complex with the Not5 subunit of the CCR4-NOT complex | | Descriptor: | 25S rRNA, 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Buschauer, R, Cheng, J, Berninghausen, O, Tesina, P, Becker, T, Beckmann, R. | | Deposit date: | 2019-10-31 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The Ccr4-Not complex monitors the translating ribosome for codon optimality.
Science, 368, 2020
|
|
5A8L
 
 | | Human eRF1 and the hCMV nascent peptide in the translation termination complex | | Descriptor: | 28S RIBOSOMAL RNA, 60S RIBOSOMAL PROTEIN L12, 60S RIBOSOMAL PROTEIN L17, ... | | Authors: | Matheisl, S, Berninghausen, O, Becker, T, Beckmann, R. | | Deposit date: | 2015-07-16 | | Release date: | 2015-12-02 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of a Human Translation Termination Complex.
Nucleic Acids Res., 43, 2015
|
|
6TNU
 
 | | Yeast 80S ribosome in complex with eIF5A and decoding A-site and P-site tRNAs. | | Descriptor: | 18S rRNA, 25S rRNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, ... | | Authors: | Buschauer, R, Cheng, J, Berninghausen, O, Tesina, P, Becker, T, Beckmann, R. | | Deposit date: | 2019-12-10 | | Release date: | 2020-04-22 | | Last modified: | 2020-04-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The Ccr4-Not complex monitors the translating ribosome for codon optimality.
Science, 368, 2020
|
|
6TMF
 
 | | Structure of an archaeal ABCE1-bound ribosomal post-splitting complex | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Kratzat, H, Becker, T, Tampe, R, Beckmann, R. | | Deposit date: | 2019-12-04 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular analysis of the ribosome recycling factor ABCE1 bound to the 30S post-splitting complex.
Embo J., 39, 2020
|
|
5A7U
 
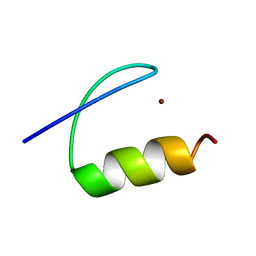 | | Single-particle cryo-EM of co-translational folded adr1 domain inside the E. coli ribosome exit tunnel. | | Descriptor: | REGULATORY PROTEIN ADR1, ZINC ION | | Authors: | Nilsson, O.B, Hedman, R, Marino, J, Wickles, S, Bischoff, L, Johansson, M, Muller-Lucks, A, Trovato, F, Puglisi, J.D, O'Brien, E, Beckmann, R, von Heijne, G. | | Deposit date: | 2015-07-10 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cotranslational Protein Folding Inside the Ribosome Exit Tunnel.
Cell Rep., 12, 2015
|
|
3ELS
 
 | | Crystal Structure of Yeast Pml1p, Residues 51-204 | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Trowitzsch, S, Weber, G, Luehrmann, R, Wahl, M.C. | | Deposit date: | 2008-09-23 | | Release date: | 2009-02-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the Pml1p subunit of the yeast precursor mRNA retention and splicing complex.
J.Mol.Biol., 385, 2009
|
|
2MK0
 
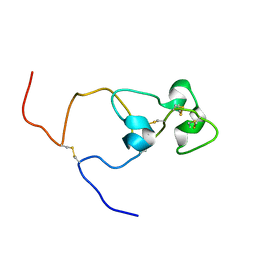 | | Structure of the PSCD4-domain of the cell wall protein pleuralin-1 from the diatom Cylindrotheca fusiformis | | Descriptor: | HEP200 protein | | Authors: | De Sanctis, S, Wenzler, M, Kroeger, N, Malloni, W.M, Sumper, M, Deutzmann, R, Zadravec, P, Brunner, E, Kremer, W, Kalbitzer, S.H.R. | | Deposit date: | 2014-01-22 | | Release date: | 2015-02-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | PSCD Domains of Pleuralin-1 from the Diatom Cylindrotheca fusiformis: NMR Structures and Interactions with Other Biosilica-Associated Proteins.
Structure, 24, 2016
|
|
4HWY
 
 | | Trypanosoma brucei procathepsin B solved from 40 fs free-electron laser pulse data by serial femtosecond X-ray crystallography | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cysteine peptidase C (CPC), beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Redecke, L, Nass, K, DePonte, D.P, White, T.A, Rehders, D, Barty, A, Stellato, F, Liang, M, Barends, T.R.M, Boutet, S, Williams, G.W, Messerschmidt, M, Seibert, M.M, Aquila, A, Arnlund, D, Bajt, S, Barth, T, Bogan, M.J, Caleman, C, Chao, T.-C, Doak, R.B, Fleckenstein, H, Frank, M, Fromme, R, Galli, L, Grotjohann, I, Hunter, M.S, Johansson, L.C, Kassemeyer, S, Katona, G, Kirian, R.A, Koopmann, R, Kupitz, C, Lomb, L, Martin, A.V, Mogk, S, Neutze, R, Shoemann, R.L, Steinbrener, J, Timneanu, N, Wang, D, Weierstall, U, Zatsepin, N.A, Spence, J.C.H, Fromme, P, Schlichting, I, Duszenko, M, Betzel, C, Chapman, H. | | Deposit date: | 2012-11-09 | | Release date: | 2012-12-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Natively inhibited Trypanosoma brucei cathepsin B structure determined by using an X-ray laser.
Science, 339, 2013
|
|
2OZB
 
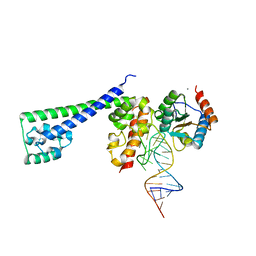 | | Structure of a human Prp31-15.5K-U4 snRNA complex | | Descriptor: | CALCIUM ION, RNA comprising the 5' Stem-Loop RNA of U4snRNA, U4/U6 small nuclear ribonucleoprotein Prp31, ... | | Authors: | Liu, S, Luehrmann, R, Wahl, M.C. | | Deposit date: | 2007-02-26 | | Release date: | 2007-03-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Binding of the human Prp31 Nop domain to a composite RNA-protein platform in U4 snRNP.
Science, 316, 2007
|
|
6R86
 
 | | Yeast Vms1-60S ribosomal subunit complex (post-state) | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Su, T, Izawa, T, Cheng, J, Yamashita, Y, Berninghausen, O, Inada, T, Neupert, W, Beckmann, R. | | Deposit date: | 2019-03-31 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and function of Vms1 and Arb1 in RQC and mitochondrial proteome homeostasis.
Nature, 570, 2019
|
|
6R87
 
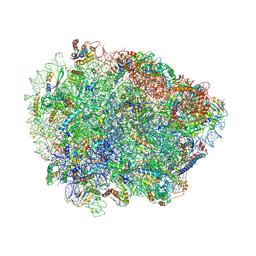 | | Yeast Vms1 (Q295L)-60S ribosomal subunit complex (pre-state without Arb1) | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Su, T, Izawa, T, Cheng, J, Yamashita, Y, Berninghausen, O, Inada, T, Neupert, W, Beckmann, R. | | Deposit date: | 2019-03-31 | | Release date: | 2019-06-26 | | Last modified: | 2019-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and function of Vms1 and Arb1 in RQC and mitochondrial proteome homeostasis.
Nature, 570, 2019
|
|
6R5Q
 
 | | Structure of XBP1u-paused ribosome nascent chain complex (post-state) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Shanmuganathan, V, Cheng, J, Berninghausen, O, Beckmann, R. | | Deposit date: | 2019-03-25 | | Release date: | 2019-07-10 | | Last modified: | 2019-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and mutational analysis of the ribosome-arresting human XBP1u.
Elife, 8, 2019
|
|
