6R1W
 
 | |
6R60
 
 | |
6R1C
 
 | |
1DNV
 
 | | PARVOVIRUS (DENSOVIRUS) FROM GALLERIA MELLONELLA | | Descriptor: | GALLERIA MELLONELLA DENSOVIRUS CAPSID PROTEIN | | Authors: | Simpson, A.A, Chipmann, P.R, Baker, T.S, Tijssen, P, Rossmann, M.G. | | Deposit date: | 1998-07-22 | | Release date: | 1999-02-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The structure of an insect parvovirus (Galleria mellonella densovirus) at 3.7 A resolution.
Structure, 6, 1998
|
|
1NCG
 
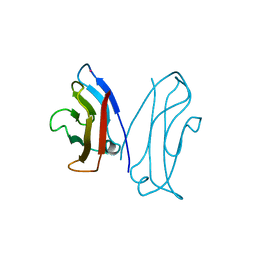 | | STRUCTURAL BASIS OF CELL-CELL ADHESION BY CADHERINS | | Descriptor: | N-CADHERIN, YTTERBIUM (III) ION | | Authors: | Shapiro, L, Fannon, A.M, Kwong, P.D, Thompson, A, Lehmann, M.S, Grubel, G, Legrand, J.-F, Als-Nielsen, J, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1995-03-23 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cell-cell adhesion by cadherins.
Nature, 374, 1995
|
|
1NCI
 
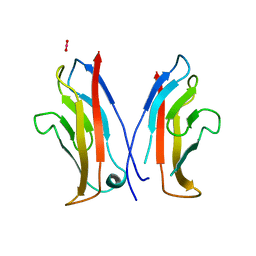 | | STRUCTURAL BASIS OF CELL-CELL ADHESION BY CADHERINS | | Descriptor: | N-CADHERIN, URANYL (VI) ION | | Authors: | Shapiro, L, Fannon, A.M, Kwong, P.D, Thompson, A, Lehmann, M.S, Grubel, G, Legrand, J.-F, Als-Nielsen, J, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1995-03-23 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cell-cell adhesion by cadherins.
Nature, 374, 1995
|
|
2BPA
 
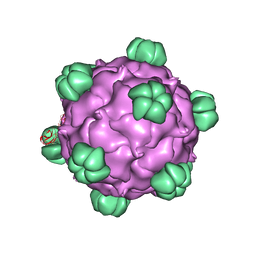 | | ATOMIC STRUCTURE OF SINGLE-STRANDED DNA BACTERIOPHAGE PHIX174 AND ITS FUNCTIONAL IMPLICATIONS | | Descriptor: | DNA (5'-D(*AP*AP*AP*AP*C)-3'), PROTEIN (SUBUNIT OF BACTERIOPHAGE PHIX174) | | Authors: | McKenna, R, Xia, D, Willingmann, P, Ilag, L.L, Krishnaswamy, S, Rossmann, M.G, Olson, N.H, Baker, T.S, Incardona, N.L. | | Deposit date: | 1991-12-03 | | Release date: | 1991-12-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Atomic structure of single-stranded DNA bacteriophage phi X174 and its functional implications.
Nature, 355, 1992
|
|
1PDL
 
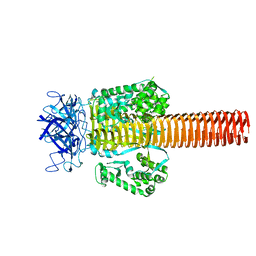 | | Fitting of gp5 in the cryoEM reconstruction of the bacteriophage T4 baseplate | | Descriptor: | Tail-associated lysozyme | | Authors: | Kostyuchenko, V.A, Leiman, P.G, Chipman, P.R, Kanamaru, S, van Raaij, M.J, Arisaka, F, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2003-05-19 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Three-dimensional structure of the bacteriophage T4 baseplate
Nat.Struct.Biol., 10, 2003
|
|
1PDJ
 
 | | Fitting of gp27 into cryoEM reconstruction of bacteriophage T4 baseplate | | Descriptor: | Baseplate structural protein Gp27 | | Authors: | Kostyuchenko, V.A, Leiman, P.G, Chipman, P.R, Kanamaru, S, van Raaij, M.J, Arisaka, F, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2003-05-19 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Three-dimensional structure of the bacteriophage T4 baseplate
Nat.Struct.Biol., 10, 2003
|
|
1PDP
 
 | | Fitting of gp9 structure into the bacteriophage T4 baseplate cryoEM reconstruction | | Descriptor: | Baseplate structural protein Gp9 | | Authors: | Kostyuchenko, V.A, Leiman, P.G, Chipman, P.R, Kanamaru, S, van Raaij, M.J, Arisaka, F, Mesyanzhinov, V.V, Rossmann, M.G. | | Deposit date: | 2003-05-19 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Three-dimensional structure of the bacteriophage T4 baseplate
Nat.Struct.Biol., 10, 2003
|
|
2N5P
 
 | | Universal base control oligonucleotide structure | | Descriptor: | DNA_(5'-D(*AP*TP*GP*GP*AP*GP*CP*TP*C)-3'), DNA_(5'-D(*GP*AP*GP*CP*TP*CP*CP*AP*T)-3') | | Authors: | Spring-Connell, A.M, Evich, M.G, Seela, F, Germann, M.W. | | Deposit date: | 2015-07-23 | | Release date: | 2016-09-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Using NMR and molecular dynamics to link structure and dynamics effects of the universal base 8-aza, 7-deaza, N8 linked adenosine analog.
Nucleic Acids Res., 44, 2016
|
|
1Z7Z
 
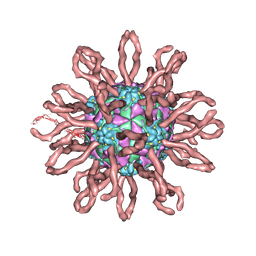 | | Cryo-em structure of human coxsackievirus A21 complexed with five domain icam-1kilifi | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Intercellular adhesion molecule-1, human coxsackievirus A21 | | Authors: | Xiao, C, Bator-Kelly, C.M, Rieder, E, Chipman, P.R, Craig, A, Kuhn, R.J, Wimmer, E, Rossmann, M.G. | | Deposit date: | 2005-03-28 | | Release date: | 2005-08-02 | | Last modified: | 2021-10-20 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | The crystal structure of coxsackievirus a21 and its interaction with icam-1.
Structure, 13, 2005
|
|
1O9W
 
 | | F17-aG lectin domain from Escherichia coli in complex with N-acetyl-glucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F17A-G FIMBRIAL ADHESIN | | Authors: | Buts, L, De Genst, E, Loris, R, Oscarson, S, Lahmann, M, Messens, J, Brosens, E, Wyns, L, Bouckaert, J, De Greve, H. | | Deposit date: | 2002-12-20 | | Release date: | 2003-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Fimbrial Adhesin F17-G of Enterotoxigenic Escherichia Coli Has an Immunoglobulin-Like Lectin Domain that Binds N-Acetylglucosamine
Mol.Microbiol., 49, 2003
|
|
2Y85
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS PHOSPHORIBOSYL ISOMERASE WITH BOUND RCDRP | | Descriptor: | 1-(O-CARBOXY-PHENYLAMINO)-1-DEOXY-D-RIBULOSE-5-PHOSPHATE, CHLORIDE ION, PHOSPHORIBOSYL ISOMERASE A, ... | | Authors: | Kuper, J, Geerlof, A, Wilmanns, M. | | Deposit date: | 2011-02-03 | | Release date: | 2011-03-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bisubstrate Specificity in Histidine/Tryptophan Biosynthesis Isomerase from Mycobacterium Tuberculosis by Active Site Metamorphosis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1NCH
 
 | | STRUCTURAL BASIS OF CELL-CELL ADHESION BY CADHERINS | | Descriptor: | N-CADHERIN, YTTERBIUM (III) ION | | Authors: | Shapiro, L, Fannon, A.M, Kwong, P.D, Thompson, A, Lehmann, M.S, Grubel, G, Legrand, J.-F, Als-Nielsen, J, Colman, D.R, Hendrickson, W.A. | | Deposit date: | 1995-03-23 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of cell-cell adhesion by cadherins.
Nature, 374, 1995
|
|
5AFG
 
 | | Structure of the Stapled Peptide Bound to Mdm2 | | Descriptor: | 1,8-DIETHYL-1,8-DIHYDRODIBENZO[3,4:7,8][1,2,3]TRIAZOLO[4',5':5,6]CYCLOOCTA[1,2-D][1,2,3]TRIAZOLE, E3 UBIQUITIN-PROTEIN LIGASE MDM2, STAPLED PEPTIDE | | Authors: | Lau, Y.H, Wu, Y, Rossmann, M, de Andrade, P, Tan, Y.S, McKenzie, G.J, Venkitaraman, A.R, Hyvonen, M, Spring, D.R. | | Deposit date: | 2015-01-22 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Double Strain-Promoted Macrocyclization for the Rapid Selection of Cell-Active Stapled Peptides.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
2IDF
 
 | | P. aeruginosa azurin N42C/M64E double mutant, BMME-linked dimer | | Descriptor: | 1-[PYRROL-1-YL-2,5-DIONE-METHOXYMETHYL]-PYRROLE-2,5-DIONE, Azurin, COPPER (II) ION, ... | | Authors: | Einsle, O, de Jongh, T.E, Hoffmann, M, Cavazzini, D, Rossi, G.L, Ubbink, M, Canters, G.W. | | Deposit date: | 2006-09-15 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Electron transfer in a crosslinked protein dimer mediated by a hydrogen-bonded network across the dimer interface
To be Published
|
|
1O9Z
 
 | | F17-aG lectin domain from Escherichia coli (ligand free) | | Descriptor: | F17-AG LECTIN DOMAIN | | Authors: | Buts, L, De Genst, E, Loris, R, Oscarson, S, Lahmann, M, Messens, J, Brosens, E, Wyns, L, Bouckaert, J, De Greve, H. | | Deposit date: | 2002-12-23 | | Release date: | 2003-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Fimbrial Adhesin F17-G of Enterotoxigenic Escherichia Coli Has an Immunoglobulin-Like Lectin Domain that Binds N-Acetylglucosamine
Mol.Microbiol., 49, 2003
|
|
5A1Q
 
 | |
6R5X
 
 | |
6R5Y
 
 | |
5ADP
 
 | | Crystal structure of the A.17 antibody FAB fragment - Light chain S35R mutant | | Descriptor: | FAB A.17 | | Authors: | Chatziefthimiou, S.D, Smirnov, I.V, Golovin, A.V, Stepanova, A.V, Peng, Y, Zolotareva, O.I, Belogurov, A.A, Ponomarenko, N.A, Blackburn, G.M, Gabibov, A.A, Lerner, R, Wilmanns, M. | | Deposit date: | 2015-08-21 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Robotic Qm/Mm-Driven Maturation of Antibody Combining Sites.
Sci.Adv., 2, 2016
|
|
4PF4
 
 | | 1.1A X-RAY STRUCTURE OF THE APO CATALYTIC DOMAIN OF DEATH-ASSOCIATED PROTEIN KINASE 1, aa 1-277 | | Descriptor: | Death-associated protein kinase 1, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Temmerman, K, Simon, B, Wilmanns, M. | | Deposit date: | 2014-04-28 | | Release date: | 2014-05-14 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | 1.1A X-RAY STRUCTURE OF THE APO CATALYTIC DOMAIN OF DEATH-ASSOCIATED PROTEIN KINASE 1, aa 1-277
To Be Published
|
|
1YE8
 
 | | Crystal Structure of THEP1 from the hyperthermophile Aquifex aeolicus | | Descriptor: | Hypothetical UPF0334 kinase-like protein AQ_1292, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Rossbach, M, Daumke, O, Klinger, C, Wittinghofer, A, Kaufmann, M. | | Deposit date: | 2004-12-28 | | Release date: | 2005-03-29 | | Last modified: | 2014-11-12 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of THEP1 from the hyperthermophile Aquifex aeolicus: a variation of the RecA fold
BMC Struct.Biol., 5, 2005
|
|
2OF6
 
 | | Structure of immature West Nile virus | | Descriptor: | envelope glycoprotein E | | Authors: | Zhang, Y, Kaufmann, B, Chipman, P.R, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2007-01-02 | | Release date: | 2007-04-03 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (24 Å) | | Cite: | Structure of immature west nile virus.
J.Virol., 81, 2007
|
|
