5OBJ
 
 | | Aurora A kinase in complex with 2-(3-fluorophenyl)quinoline-4-carboxylic acid and ATP | | Descriptor: | 2-(3-fluorophenyl)quinoline-4-carboxylic acid, ADENOSINE-5'-TRIPHOSPHATE, Aurora kinase A, ... | | Authors: | Rossmann, M, Janecek, M, Hyvonen, M. | | Deposit date: | 2017-06-28 | | Release date: | 2017-08-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Computationally-guided optimization of small-molecule inhibitors of the Aurora A kinase-TPX2 protein-protein interaction.
Chem. Commun. (Camb.), 53, 2017
|
|
5N67
 
 | |
5N68
 
 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9m | | Descriptor: | 2-(4-morpholin-4-ylphenyl)-~{N}4-(2-phenylethyl)quinazoline-4,7-diamine, Mitogen-activated protein kinase 14 | | Authors: | Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
5N63
 
 | | Crystal Structure of p38alpha in Complex with Lipid Pocket Ligand 9c | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}4-[(4-fluorophenyl)methyl]-2-phenyl-quinazoline-4,7-diamine | | Authors: | Buehrmann, M, Mueller, M.P, Rauh, D. | | Deposit date: | 2017-02-14 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design, synthesis and crystallization of 2-arylquinazolines as lipid pocket ligands of p38 alpha MAPK.
PLoS ONE, 12, 2017
|
|
3JS8
 
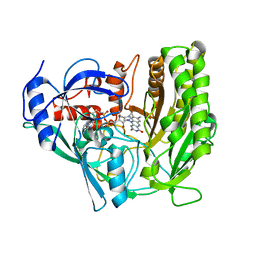 | | Solvent-stable cholesterol oxidase | | Descriptor: | Cholesterol oxidase, FLAVIN-ADENINE DINUCLEOTIDE, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Sagermann, M, Ohtaki, A, Newton, K, Doukyu, N. | | Deposit date: | 2009-09-09 | | Release date: | 2010-02-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural characterization of the organic solvent-stable cholesterol oxidase from Chromobacterium sp. DS-1.
J.Struct.Biol., 170, 2010
|
|
5ML5
 
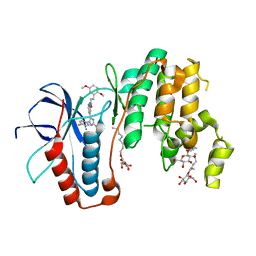 | | Human p38alpha MAPK in complex with imidazolyl pyridine inhibitor 11b | | Descriptor: | 3-(2,5-dimethoxyphenyl)-~{N}-[4-[4-(4-fluorophenyl)-2-methylsulfanyl-1~{H}-imidazol-5-yl]pyridin-2-yl]propanamide, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Buehrmann, M, Rauh, D. | | Deposit date: | 2016-12-06 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimized 4,5-Diarylimidazoles as Potent/Selective Inhibitors of Protein Kinase CK1 delta and Their Structural Relation to p38 alpha MAPK.
Molecules, 22, 2017
|
|
3HQ8
 
 | | CcpA from G. sulfurreducens S134P/V135K variant | | Descriptor: | CALCIUM ION, Cytochrome c551 peroxidase, IMIDAZOLE, ... | | Authors: | Hoffmann, M, Seidel, J, Einsle, O. | | Deposit date: | 2009-06-05 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CcpA from Geobacter sulfurreducens is a basic di-heme cytochrome c peroxidase.
J.Mol.Biol., 393, 2009
|
|
5LOS
 
 | | Piriformospora indica PIIN_05872 | | Descriptor: | PIIN_05872, SULFATE ION, ZINC ION | | Authors: | Hartmann, M.D, Albrecht, R, Martin, J, Lupas, A.N. | | Deposit date: | 2016-08-09 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A secreted fungal histidine- and alanine-rich protein regulates metal ion homeostasis and oxidative stress.
New Phytol., 227, 2020
|
|
3HQ7
 
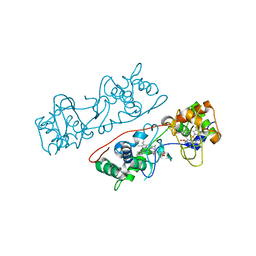 | | CcpA from G. sulfurreducens, G94K/K97Q/R100I variant | | Descriptor: | CALCIUM ION, Cytochrome c551 peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hoffmann, M, Seidel, J, Einsle, O. | | Deposit date: | 2009-06-05 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | CcpA from Geobacter sulfurreducens is a basic di-heme cytochrome c peroxidase.
J.Mol.Biol., 393, 2009
|
|
3GFH
 
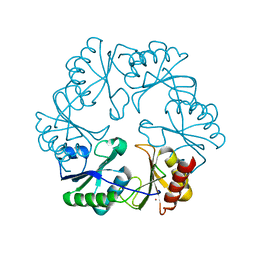 | |
5OMG
 
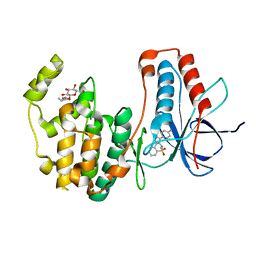 | | p38alpha in complex with pyrazolobenzothiazine inhibitor COXP4M12 | | Descriptor: | 3-(4-fluorophenyl)-4-methyl-1~{H}-pyrazolo[4,3-c][1,2]benzothiazine 5,5-dioxide, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Buehrmann, M, Rauh, D. | | Deposit date: | 2017-07-31 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Co-crystal structure determination and cellular evaluation of 1,4-dihydropyrazolo[4,3-c] [1,2] benzothiazine 5,5-dioxide p38 alpha MAPK inhibitors.
Biochem.Biophys.Res.Commun., 511, 2019
|
|
5OMH
 
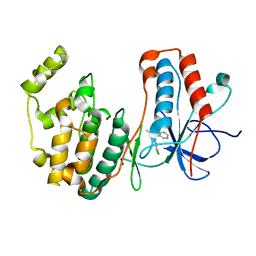 | | p38alpha in complex with pyrazolobenzothiazine inhibitor COXH11 | | Descriptor: | 1-(3-chlorophenyl)-3-methyl-4~{H}-pyrazolo[4,3-c][1,2]benzothiazine 5,5-dioxide, Mitogen-activated protein kinase 14 | | Authors: | Buehrmann, M, Rauh, D. | | Deposit date: | 2017-07-31 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Co-crystal structure determination and cellular evaluation of 1,4-dihydropyrazolo[4,3-c] [1,2] benzothiazine 5,5-dioxide p38 alpha MAPK inhibitors.
Biochem.Biophys.Res.Commun., 511, 2019
|
|
5TBE
 
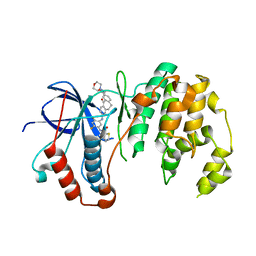 | | Human p38alpha MAP Kinase in Complex with Dibenzosuberone Compound 2 | | Descriptor: | Mitogen-activated protein kinase 14, ~{N}-[2,4-bis(fluoranyl)-5-[[9-(2-morpholin-4-ylethylcarbamoyl)-11-oxidanylidene-5,6-dihydrodibenzo[1,2-~{d}:1',2'-~{f}][7]annulen-3-yl]amino]phenyl]thiophene-2-carboxamide | | Authors: | Buehrmann, M, Rauh, D. | | Deposit date: | 2016-09-12 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Optimized Target Residence Time: Type I1/2 Inhibitors for p38 alpha MAP Kinase with Improved Binding Kinetics through Direct Interaction with the R-Spine.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5VU2
 
 | |
2WQ2
 
 | |
1TUC
 
 | |
2WPR
 
 | | Salmonella enterica SadA 483-523 fused to GCN4 adaptors (SadAK3b-V1, out-of-register fusion) | | Descriptor: | CHLORIDE ION, TRIMERIC AUTOTRANSPORTER ADHESIN FRAGMENT | | Authors: | Hartmann, M.D, Ridderbusch, O, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2009-08-09 | | Release date: | 2009-11-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A Coiled-Coil Motif that Sequesters Ions to the Hydrophobic Core.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2WQ3
 
 | |
5O8U
 
 | |
5O8V
 
 | |
8V4G
 
 | | X-ray structure of the NADP-dependent reductase from Campylobacter jejuni responsible for the synthesis of CDP-glucitol in the presence of CDP and NADP | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CYTIDINE-5'-DIPHOSPHATE, ... | | Authors: | Schumann, M.E, Thoden, J.B, Holden, H.M, Raushel, F.M. | | Deposit date: | 2023-11-29 | | Release date: | 2023-12-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biosynthesis of Cytidine Diphosphate-6-d-Glucitol for the Capsular Polysaccharides of Campylobacter jejuni.
Biochemistry, 63, 2024
|
|
3ZMF
 
 | | Salmonella enterica SadA 303-358 fused to GCN4 adaptors (SadAK2) | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, PUTATIVE INNER MEMBRANE PROTEIN | | Authors: | Hartmann, M.D, Hernandez Alvarez, B, Albrecht, R, Lupas, A.N. | | Deposit date: | 2013-02-08 | | Release date: | 2013-02-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A New Expression System for Protein Crystallization Using Trimeric Coiled-Coil Adaptors.
Protein Eng.Des.Sel., 21, 2008
|
|
7LJX
 
 | | Oxidized rat cytochrome c mutant (K53Q) | | Descriptor: | Cytochrome c, somatic, HEME C, ... | | Authors: | Huttemann, M, Edwards, B.F.P, Brunzelle, J.S, Vaishnav, A. | | Deposit date: | 2021-02-01 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Lysine 53 Acetylation of Cytochrome c in Prostate Cancer: Warburg Metabolism and Evasion of Apoptosis.
Cells, 10, 2021
|
|
4UIB
 
 | | Crystal structure of 3p in complex with tafCPB | | Descriptor: | (2S)-6-AMINO-2-[[(1R)-1-(CYCLOHEXYLMETHYL)-2-OXO-2-[[(2S)-1,7,7-TRIMETHYLNORBORNAN-2-YL]AMINO]ETHYL]ARBAMOYLAMINO]HEXANOIC ACID, ACETATE ION, CARBOXYPEPTIDASE B, ... | | Authors: | Halland, N, Broenstrup, M, Czech, J, Czechtizky, W, Evers, A, Follmann, M, Kohlmann, M, Schiell, M, Kurz, M, Schreuder, H.A, Kallus, C. | | Deposit date: | 2015-03-27 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Novel Small Molecule Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (Tafia) from Natural Product Anabaenopeptin.
J.Med.Chem., 58, 2015
|
|
4UIA
 
 | | Crystal structure of 3a in complex with tafCPB | | Descriptor: | (2S)-6-azanyl-2-[[(2R)-3-cyclohexyl-1-(3-methylbutylamino)-1-oxidanylidene-propan-2-yl]carbamoylamino]hexanoic acid, CARBOXYPEPTIDASE B, ZINC ION | | Authors: | Halland, N, Broenstrup, M, Czech, J, Czechtizky, W, Evers, A, Follmann, M, Kohlmann, M, Schiell, M, Kurz, M, Schreuder, H.A, Kallus, C. | | Deposit date: | 2015-03-27 | | Release date: | 2015-06-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Novel Small Molecule Inhibitors of Activated Thrombin Activatable Fibrinolysis Inhibitor (Tafia) from Natural Product Anabaenopeptin.
J.Med.Chem., 58, 2015
|
|
