5LYX
 
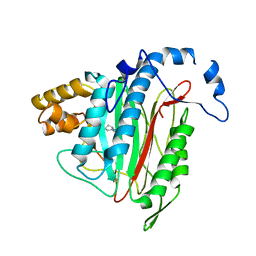 | | CRYSTAL STRUCTURE OF HUMAN METHIONINE AMINOPEPTIDASE-2 IN COMPLEX; WITH AN INHIBITOR 5-((R)-1-[1,2,4]Triazolo[1,5-a]pyrimidin-7-yl-pyrrolidin-2-ylmethoxy)-isoquinoline | | Descriptor: | 5-[[(2~{R})-1-([1,2,4]triazolo[1,5-a]pyrimidin-7-yl)pyrrolidin-2-yl]methoxy]isoquinoline, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Musil, D, Heinrich, T, Lehmann, M. | | Deposit date: | 2016-09-29 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel reversible methionine aminopeptidase-2 (MetAP-2) inhibitors based on purine and related bicyclic templates.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
3ZYH
 
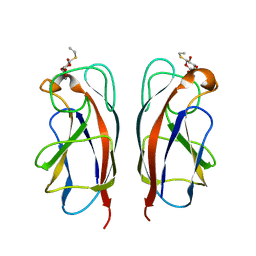 | | CRYSTAL STRUCTURE OF PA-IL LECTIN COMPLEXED WITH GALBG0 AT 1.5 A RESOLUTION | | Descriptor: | 3-(beta-D-galactopyranosylthio)propanoic acid, CALCIUM ION, PA-I galactophilic lectin | | Authors: | Kadam, R.U, Bergmann, M, Hurley, M, Garg, D, Cacciarini, M, Swiderska, M.A, Nativi, C, Sattler, M, Smyth, A.R, Williams, P, Camara, M, Stocker, A, Darbre, T, Reymond, J.-L. | | Deposit date: | 2011-08-23 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | A Glycopeptide Dendrimer Inhibitor of the Galactose Specific Lectin Leca and of Pseudomonas Aeruginosa Biofilms
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3ZYB
 
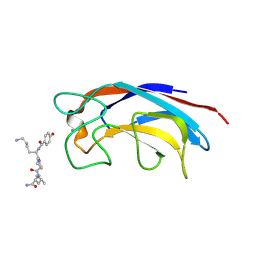 | | CRYSTAL STRUCTURE OF PA-IL LECTIN COMPLEXED WITH GALAG0 AT 2.3 A RESOLUTION | | Descriptor: | CALCIUM ION, GALA-LYS-PRO-LEUNH2, P-HYDROXYBENZOIC ACID, ... | | Authors: | Kadam, R.U, Bergmann, M, Hurley, M, Garg, D, Cacciarini, M, Swiderska, M.A, Nativi, C, Sattler, M, Smyth, A.R, Williams, P, Camara, M, Stocker, A, Darbre, T, Reymond, J.-L. | | Deposit date: | 2011-08-19 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A Glycopeptide Dendrimer Inhibitor of the Galactose-Specific Lectin Leca and of Pseudomonas Aeruginosa Biofilms.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
3ZYF
 
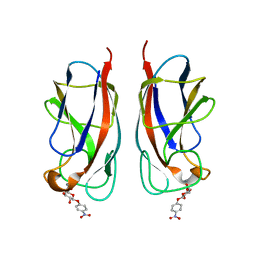 | | CRYSTAL STRUCTURE OF PA-IL LECTIN COMPLEXED WITH NPG AT 1.9 A RESOLUTION | | Descriptor: | 4-nitrophenyl beta-D-galactopyranoside, CALCIUM ION, PA-I GALACTOPHILIC LECTIN | | Authors: | Kadam, R.U, Bergmann, M, Hurley, M, Garg, D, Cacciarini, M, Swiderska, M.A, Nativi, C, Sattler, M, Smyth, A.R, Williams, P, Camara, M, Stocker, A, Darbre, T, Reymond, J.-L. | | Deposit date: | 2011-08-22 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.944 Å) | | Cite: | A Glycopeptide Dendrimer Inhibitor of the Galactose-Specific Lectin Leca and of Pseudomonas Aeruginosa Biofilms.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
2C0M
 
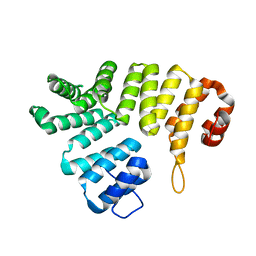 | |
3ZNJ
 
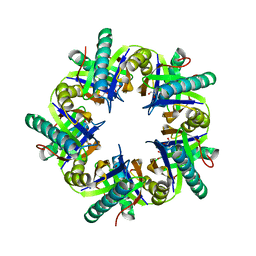 | | Crystal structure of unliganded ClcF from R.opacus 1CP in crystal form 1. | | Descriptor: | 1,2-ETHANEDIOL, 5-CHLOROMUCONOLACTONE DEHALOGENASE, CHLORIDE ION | | Authors: | Roth, C, Groening, J.A.D, Kaschabek, S.R, Schloemann, M, Straeter, N. | | Deposit date: | 2013-02-14 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Catalytic Mechanism of Chloromuconolactone Dehalogenase Clcf from Rhodococcus Opacus 1Cp.
Mol.Microbiol., 88, 2013
|
|
2CGH
 
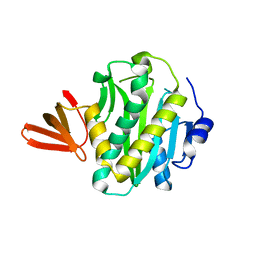 | |
4LGL
 
 | | Crystal Structure of Glycine Decarboxylase P-protein from Synechocystis sp. PCC 6803, apo form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Glycine dehydrogenase [decarboxylating] | | Authors: | Hasse, D, Andersson, E, Carlsson, G, Masloboy, A, Hagemann, M, Bauwe, H, Andersson, I. | | Deposit date: | 2013-06-28 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.0004 Å) | | Cite: | Structure of the Homodimeric Glycine Decarboxylase P-protein from Synechocystis sp. PCC 6803 Suggests a Mechanism for Redox Regulation.
J.Biol.Chem., 288, 2013
|
|
3SHS
 
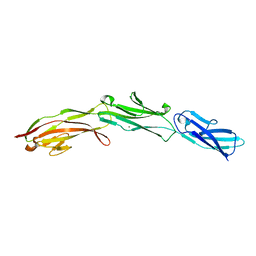 | | Three N-terminal domains of the bacteriophage RB49 Highly Immunogenic Outer Capsid protein (Hoc) | | Descriptor: | Hoc head outer capsid protein, MAGNESIUM ION | | Authors: | Fokine, A, Islam, M.Z, Zhang, Z, Bowman, V.D, Rao, V.B, Rossmann, M.G. | | Deposit date: | 2011-06-16 | | Release date: | 2011-06-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Structure of the three N-terminal immunoglobulin domains of the highly immunogenic outer capsid protein from a T4-like bacteriophage.
J.Virol., 85, 2011
|
|
5NN1
 
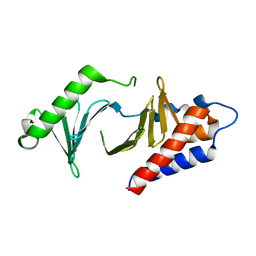 | |
2VCD
 
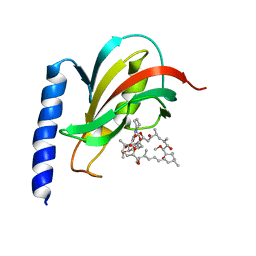 | | Solution structure of the FKBP-domain of Legionella pneumophila Mip in complex with rapamycin | | Descriptor: | Outer membrane protein MIP, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Ceymann, A, Horstmann, M, Ehses, P, Schweimer, K, Paschke, A.-K, Fischer, G, Roesch, P, Faber, C. | | Deposit date: | 2007-09-20 | | Release date: | 2008-09-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Legionella pneumophila Mip-rapamycin complex.
BMC Struct. Biol., 8, 2008
|
|
2B6B
 
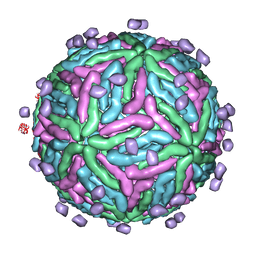 | | Cryo EM structure of Dengue complexed with CRD of DC-SIGN | | Descriptor: | CD209 antigen, envelope glycoprotein | | Authors: | Pokidysheva, E, Zhang, Y, Battisti, A.J, Bator-Kelly, C.M, Chipman, P.R, Gregorio, G, Hendrickson, W.A, Kuhn, R.J, Rossmann, M.G. | | Deposit date: | 2005-09-30 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Cryo-EM reconstruction of dengue virus in complex with the carbohydrate recognition domain of DC-SIGN
Cell(Cambridge,Mass.), 124, 2006
|
|
1FYN
 
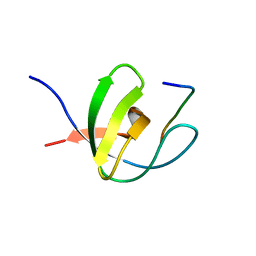 | | PHOSPHOTRANSFERASE | | Descriptor: | 3BP-2, PHOSPHOTRANSFERASE FYN | | Authors: | Musacchio, A, Saraste, M, Wilmanns, M. | | Deposit date: | 1995-05-17 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High-resolution crystal structures of tyrosine kinase SH3 domains complexed with proline-rich peptides.
Nat.Struct.Biol., 1, 1994
|
|
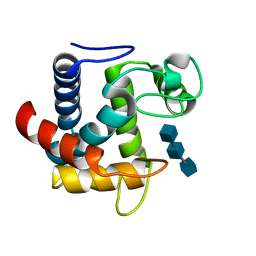
3CSZ
 
 | |
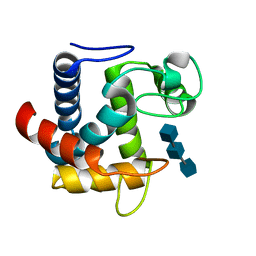
3CT5
 
 | |
2W84
 
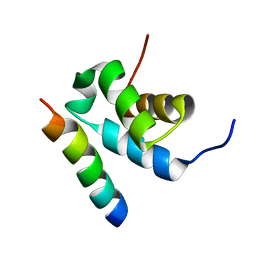 | | Structure of Pex14 in complex with Pex5 | | Descriptor: | PEROXISOMAL MEMBRANE PROTEIN PEX14, PEROXISOMAL TARGETING SIGNAL 1 RECEPTOR | | Authors: | Neufeld, C, Filipp, F.V, Simon, B, Neuhaus, A, Schueller, N, David, C, Kooshapur, H, Madl, T, Erdmann, R, Schliebs, W, Wilmanns, M, Sattler, M. | | Deposit date: | 2009-01-09 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for competitive interactions of Pex14 with the import receptors Pex5 and Pex19.
EMBO J., 28, 2009
|
|
2W85
 
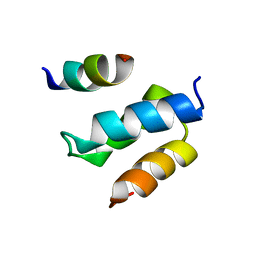 | | Structure of Pex14 in complex with Pex19 | | Descriptor: | PEROXIN-19, PEROXISOMAL MEMBRANE ANCHOR PROTEIN PEX14 | | Authors: | Neufeld, C, Filipp, F.V, Simon, B, Neuhaus, A, Schueller, N, David, C, Kooshapur, H, Madl, T, Erdmann, R, Schliebs, W, Wilmanns, M, Sattler, M. | | Deposit date: | 2009-01-09 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Competitive Interactions of Pex14 with the Import Receptors Pex5 and Pex19.
Embo J., 28, 2009
|
|
2AO7
 
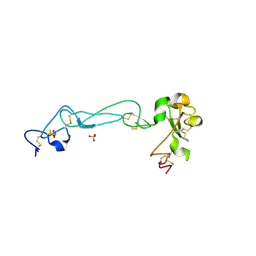 | | Adam10 Disintegrin and cysteine- rich domain | | Descriptor: | ADAM 10, SULFATE ION | | Authors: | Janes, P.W, Saha, N, Barton, W.A, Kolev, M.V, Wimmer-Kleikamp, S.H, Nievergall, E, Blobel, C.P, Himanen, J.-P, Lackmann, M, Nikolov, D.B. | | Deposit date: | 2005-08-12 | | Release date: | 2006-08-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Adam meets Eph: an ADAM substrate recognition module acts as a molecular switch for ephrin cleavage in trans.
Cell(Cambridge,Mass.), 123, 2005
|
|
3STJ
 
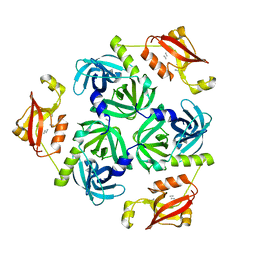 | | Crystal structure of the protease + PDZ1 domain of DegQ from Escherichia coli | | Descriptor: | Protease degQ, peptide (UNK) | | Authors: | Sawa, J, Malet, H, Krojer, T, Canellas, F, Ehrmann, M, Clausen, T. | | Deposit date: | 2011-07-11 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular adaptation of the DegQ protease to exert protein quality control in the bacterial cell envelope.
J.Biol.Chem., 286, 2011
|
|
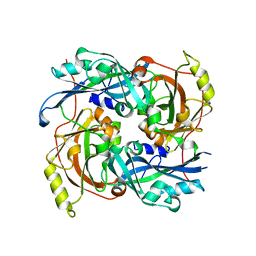
4G1G
 
 | |
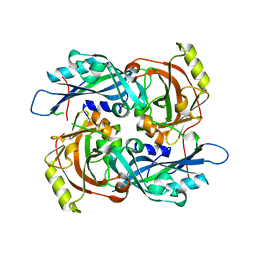
4G1L
 
 | |
1SOT
 
 | | Crystal Structure of the DegS stress sensor | | Descriptor: | Protease degS | | Authors: | Wilken, C, Kitzing, K, Kurzbauer, R, Ehrmann, M, Clausen, T. | | Deposit date: | 2004-03-15 | | Release date: | 2004-06-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease
Cell(Cambridge,Mass.), 117, 2004
|
|
4JPP
 
 | | Bacteriophage phiX174 H protein residues 143-282 | | Descriptor: | Minor spike protein H | | Authors: | Sun, L, Young, L.N, Boudko, S.B, Fokine, A, Zhang, X, Rossmann, M.G, Fane, B.A. | | Deposit date: | 2013-03-19 | | Release date: | 2013-12-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Icosahedral bacteriophage Phi X174 forms a tail for DNA transport during infection.
Nature, 505, 2014
|
|
4HBO
 
 | |
3CSQ
 
 | |
