4AE3
 
 | | Crystal structure of ammosamide 272:myosin-2 motor domain complex | | Descriptor: | 1,2-ETHANEDIOL, ADP ORTHOVANADATE, AMMOSAMIDE 272, ... | | Authors: | Chinthalapudi, K, Heissler, S.M, Fenical, W, Manstein, D.J. | | Deposit date: | 2012-01-05 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Ammosamide Mediated Myosin Motor Activity Inhibition
To be Published
|
|
433D
 
 | |
1DML
 
 | | CRYSTAL STRUCTURE OF HERPES SIMPLEX UL42 BOUND TO THE C-TERMINUS OF HSV POL | | Descriptor: | DNA POLYMERASE, DNA POLYMERASE PROCESSIVITY FACTOR | | Authors: | Zuccola, H.J, Filman, D.J, Coen, D.M, Hogle, J.M. | | Deposit date: | 1999-12-14 | | Release date: | 2000-03-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of an unusual processivity factor, herpes simplex virus UL42, bound to the C terminus of its cognate polymerase.
Mol.Cell, 5, 2000
|
|
5MU9
 
 | | MOA-E-64 complex | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Agglutinin, ... | | Authors: | Cordara, G, Manna, D, Krengel, U. | | Deposit date: | 2017-01-12 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Family of Papain-Like Fungal Chimerolectins with Distinct Ca(2+)-Dependent Activation Mechanism.
Biochemistry, 56, 2017
|
|
6TSQ
 
 | | Marasmius oreades agglutinin (MOA) activated by manganese (II) | | Descriptor: | 1,2-ETHANEDIOL, Agglutinin, CALCIUM ION, ... | | Authors: | Cordara, G, Manna, D, Krengel, U. | | Deposit date: | 2019-12-21 | | Release date: | 2020-07-29 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of MOA in complex with a peptide fragment: A protease caught in flagranti .
Curr Res Struct Biol, 2, 2020
|
|
2X9H
 
 | | CRYSTAL STRUCTURE OF MYOSIN-2 MOTOR DOMAIN IN COMPLEX WITH ADP- METAVANADATE AND PENTACHLOROCARBAZOLE | | Descriptor: | 2,3,4,6,8-PENTACHLORO-9H-CARBAZOL-1-OL, ADP METAVANADATE, MAGNESIUM ION, ... | | Authors: | Selvadurai, J, Kirst, J, Knoelker, H.J, Manstein, D.J. | | Deposit date: | 2010-03-19 | | Release date: | 2011-05-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Myosin-2 Motor Domain in Complex with Adp-Metavanadate and Pentachlorocarbazole
To be Published
|
|
3ZJE
 
 | | A20 OTU domain in reversibly oxidised (SOH) state | | Descriptor: | 1,2-ETHANEDIOL, A20P50, CHLORIDE ION | | Authors: | Kulathu, Y, Garcia, F.J, Mevissen, T.E.T, Busch, M, Arnaudo, N, Carroll, K.S, Barford, D, Komander, D. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Regulation of A20 and Other Otu Deubiquitinases by Reversible Oxidation
Nat.Commun., 4, 2013
|
|
1GIA
 
 | |
2VZM
 
 | |
2WNY
 
 | | Structure of Mth689, a DUF54 protein from Methanothermobacter thermautotrophicus | | Descriptor: | CHLORIDE ION, CONSERVED PROTEIN MTH689, NICKEL (II) ION | | Authors: | Ng, C.L, Waterman, D.G, Lebedev, A.A, Smits, C, Ortiz-Lombardia, M, Antson, A.A. | | Deposit date: | 2009-07-21 | | Release date: | 2010-09-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Mth689, a Duf54 Protein from Methanothermobacter Thermautotrophicus
To be Published
|
|
1G6Q
 
 | | CRYSTAL STRUCTURE OF YEAST ARGININE METHYLTRANSFERASE, HMT1 | | Descriptor: | HNRNP ARGININE N-METHYLTRANSFERASE | | Authors: | Weiss, V.H, McBride, A.E, Soriano, M.A, Filman, D.J, Silver, P.A, Hogle, J.M. | | Deposit date: | 2000-11-07 | | Release date: | 2000-12-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure and oligomerization of the yeast arginine methyltransferase, Hmt1.
Nat.Struct.Biol., 7, 2000
|
|
3DDF
 
 | | GOLGI MANNOSIDASE II complex with (3R,4R,5R)-3,4-Dihydroxy-5-({[(1R)-2-hydroxy-1 phenylethyl]amino}methyl) pyrrolidin-2-one | | Descriptor: | (3R,4R,5R)-3,4-dihydroxy-5-({[(1R)-2-hydroxy-1-phenylethyl]amino}methyl)pyrrolidin-2-one, (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ... | | Authors: | Kuntz, D.A, Rose, D.R, Hoffman, D. | | Deposit date: | 2008-06-05 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Functionalized pyrrolidine inhibitors of human type II alpha-mannosidases as anti-cancer agents: optimizing the fit to the active site
Bioorg.Med.Chem., 16, 2008
|
|
4KRA
 
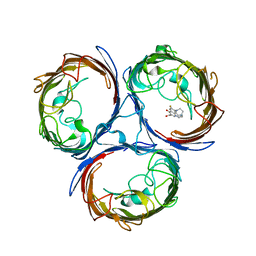 | |
3DDG
 
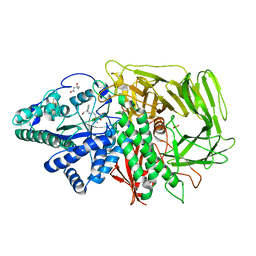 | | GOLGI MANNOSIDASE II complex with (3R,4R,5R)-3,4-Dihydroxy-5-({[(1R)-2-hydroxy-1 phenylethyl]amino}methyl) methylpyrrolidin-2-one | | Descriptor: | (3R,4R,5R)-3,4-dihydroxy-5-({[(1R)-2-hydroxy-1-phenylethyl]amino}methyl)-1-methylpyrrolidin-2-one, (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R, Hoffman, D. | | Deposit date: | 2008-06-05 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Functionalized pyrrolidine inhibitors of human type II alpha-mannosidases as anti-cancer agents: optimizing the fit to the active site
Bioorg.Med.Chem., 16, 2008
|
|
4MKI
 
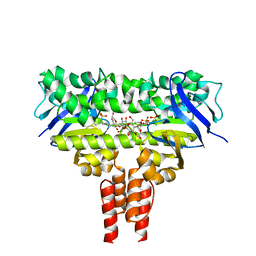 | | Cobalt transporter ATP-binding subunit | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Energy-coupling factor transporter ATP-binding protein EcfA2, SULFATE ION | | Authors: | Yu, Y, Zhang, L, Chai, C.L, Heymann, D. | | Deposit date: | 2013-09-05 | | Release date: | 2013-10-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for a homodimeric ATPase subunit of an ECF transporter
Protein Cell, 4, 2013
|
|
1G3J
 
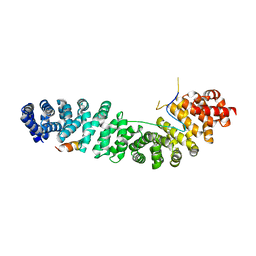 | | CRYSTAL STRUCTURE OF THE XTCF3-CBD/BETA-CATENIN ARMADILLO REPEAT COMPLEX | | Descriptor: | BETA-CATENIN ARMADILLO REPEAT REGION, TCF3-CBD (CATENIN BINDING DOMAIN) | | Authors: | Graham, T.A, Weaver, C, Mao, F, Kimelman, D, Xu, W. | | Deposit date: | 2000-10-24 | | Release date: | 2000-12-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a beta-catenin/Tcf complex.
Cell(Cambridge,Mass.), 103, 2000
|
|
1HFH
 
 | | SOLUTION STRUCTURE OF A PAIR OF COMPLEMENT MODULES BY NUCLEAR MAGNETIC RESONANCE | | Descriptor: | FACTOR H, 15TH AND 16TH C-MODULE PAIR | | Authors: | Barlow, P.N, Steinkasserer, A, Norman, D.G, Kieffer, B, Wiles, A.P, Sim, R.B, Campbell, I.D. | | Deposit date: | 1993-02-23 | | Release date: | 1993-07-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a pair of complement modules by nuclear magnetic resonance.
J.Mol.Biol., 232, 1993
|
|
1HFI
 
 | | SOLUTION STRUCTURE OF A PAIR OF COMPLEMENT MODULES BY NUCLEAR MAGNETIC RESONANCE | | Descriptor: | FACTOR H, 15TH C-MODULE PAIR | | Authors: | Barlow, P.N, Steinkasserer, A, Norman, D.G, Kieffer, B, Wiles, A.P, Sim, R.B, Campbell, I.D. | | Deposit date: | 1993-02-23 | | Release date: | 1993-07-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a pair of complement modules by nuclear magnetic resonance.
J.Mol.Biol., 232, 1993
|
|
1F1C
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME C549 | | Descriptor: | CYTOCHROME C549, HEME C | | Authors: | Kerfeld, C.A, Sawaya, M.R, Yeates, T.O, Krogmann, D.W. | | Deposit date: | 2000-05-18 | | Release date: | 2001-08-08 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of cytochrome c-549 and cytochrome c6 from the cyanobacterium Arthrospira maxima.
Biochemistry, 40, 2001
|
|
1F27
 
 | | CRYSTAL STRUCTURE OF A BIOTIN-BINDING RNA PSEUDOKNOT | | Descriptor: | BIOTIN, MAGNESIUM ION, RNA (5'-R(*AP*AP*AP*AP*AP*GP*UP*CP*CP*UP*C)-3'), ... | | Authors: | Nix, J, Sussman, D, Wilson, C. | | Deposit date: | 2000-05-23 | | Release date: | 2000-06-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The 1.3 A crystal structure of a biotin-binding pseudoknot and the basis for RNA molecular recognition.
J.Mol.Biol., 296, 2000
|
|
1HOY
 
 | | NMR STRUCTURE OF THE COMPLEX BETWEEN A-BUNGAROTOXIN AND A MIMOTOPE OF THE NICOTINIC ACETYLCHOLINE RECEPTOR | | Descriptor: | LONG NEUROTOXIN 1, MIMOTOPE OF THE NICOTINIC ACETYLCHOLINE RECEPTOR | | Authors: | Scarselli, M, Spiga, O, Ciutti, A, Bracci, L, Lelli, B, Lozzi, L, Calamandrei, D, Bernini, A, Di Maro, D, Niccolai, N, Neri, P. | | Deposit date: | 2000-12-12 | | Release date: | 2000-12-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure of alpha-bungarotoxin free and bound to a mimotope of the nicotinic acetylcholine receptor.
Biochemistry, 41, 2002
|
|
1HSN
 
 | | THE STRUCTURE OF THE HMG BOX AND ITS INTERACTION WITH DNA | | Descriptor: | BETA-MERCAPTOETHANOL, HIGH MOBILITY GROUP PROTEIN 1 | | Authors: | Read, C.M, Cary, P.D, Crane-Robinson, C, Driscoll, P.C, Carillo, M.O.M, Norman, D.G. | | Deposit date: | 1994-11-17 | | Release date: | 1995-02-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The Structure of the Hmg Box and its Interaction with DNA
To be Published
|
|
8STN
 
 | | Crystal structure of KRAS-G12D/G75A mutant, GDP-bound | | Descriptor: | CHLORIDE ION, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Tran, T.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2023-05-10 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Allosteric Regulation of Switch-II Domain Controls KRAS Oncogenicity.
Cancer Res., 83, 2023
|
|
8STM
 
 | | Crystal structure of KRAS-G75A mutant, GDP-bound | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Tran, T.H, Dharmaiah, S, Simanshu, D.K. | | Deposit date: | 2023-05-10 | | Release date: | 2023-08-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Allosteric Regulation of Switch-II Domain Controls KRAS Oncogenicity.
Cancer Res., 83, 2023
|
|
3EG5
 
 | | Crystal structure of MDIA1-TSH GBD-FH3 in complex with CDC42-GMPPNP | | Descriptor: | Cell division control protein 42 homolog, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Lammers, M, Meyer, S, Kuehlmann, D, Wittinghofer, A. | | Deposit date: | 2008-09-10 | | Release date: | 2008-10-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Specificity of Interactions between mDia Isoforms and Rho Proteins
J.Biol.Chem., 283, 2008
|
|
