6VC8
 
 | | Crystal structure of wild-type KRAS4b(1-169) in complex with GMPPNP and Mg ion | | Descriptor: | GTPase KRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | Authors: | Tran, T.H, Davies, D.R, Edwards, T.E, Simanshu, D.K. | | Deposit date: | 2019-12-20 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Machine learning-driven multiscale modeling reveals lipid-dependent dynamics of RAS signaling proteins.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6PVG
 
 | | Crystal structure of ligand free PhqK | | Descriptor: | FAD monooxygenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Fraley, A.E, Smith, J.L, Sherman, D.H. | | Deposit date: | 2019-07-20 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.709 Å) | | Cite: | Molecular Basis for Spirocycle Formation in the Paraherquamide Biosynthetic Pathway.
J.Am.Chem.Soc., 142, 2020
|
|
6Q0B
 
 | | Poliovirus (Type 1 Mahoney), receptor-catalysed 135S particle incubated with anti-VP1 mAb at RT for 1 hr | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Hogle, J.M, Filman, D.J, Shah, P.N.M. | | Deposit date: | 2019-08-01 | | Release date: | 2020-08-05 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures reveal two distinct conformational states in a picornavirus cell entry intermediate.
Plos Pathog., 16, 2020
|
|
1TB0
 
 | | Effect of Shuttle Location and pH Environment on H+ Transfer in Human Carbonic Anhydrase II | | Descriptor: | CHLORIDE ION, Carbonic anhydrase II, ZINC ION | | Authors: | Fisher, Z, Hernandez Prada, J.A, Tu, C, Duda, D, Yoshioka, C, An, H, Govindasamy, L, Silverman, D.N, McKenna, R. | | Deposit date: | 2004-05-19 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and kinetic characterization of active-site histidine as a proton shuttle in catalysis by human carbonic anhydrase II.
Biochemistry, 44, 2005
|
|
7ODZ
 
 | | Dictyostelium discoideum dye decolorizing peroxidase DyPA in complex with an activated form of oxygen and veratryl alcohol | | Descriptor: | 1,2-ETHANEDIOL, DyPA, OXYGEN MOLECULE, ... | | Authors: | Rai, A, Fedorov, R, Manstein, D.J. | | Deposit date: | 2021-04-30 | | Release date: | 2021-07-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Biochemical Characterization of a Dye-Decolorizing Peroxidase from Dictyostelium discoideum .
Int J Mol Sci, 22, 2021
|
|
1TE3
 
 | | Effect of Shuttle Location and pH Environment on H+ Transfer in Human Carbonic Anhydrase II | | Descriptor: | CHLORIDE ION, Carbonic anhydrase II, ZINC ION | | Authors: | Fisher, Z, Hernandez Prada, J.A, Tu, C.K, Duda, D, Yoshioka, C, An, H, Govindasamy, L, Silverman, D.N, McKenna, R. | | Deposit date: | 2004-05-24 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Kinetic Characterization of Active-Site Histidine as a Proton Shuttle in Catalysis by Human Carbonic Anhydrase II
Biochemistry, 44, 2005
|
|
1TEQ
 
 | | Effect of Shuttle Location and pH Environment on H+ Transfer in Human Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase II, HYDROXIDE ION, ZINC ION | | Authors: | Fisher, Z, Hernandez Prada, J.A, Tu, C.K, Duda, D, Yoshioka, C, An, H, Govindasamy, L, Silverman, D.N, McKenna, R. | | Deposit date: | 2004-05-25 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Kinetic Characterization of Active-Site Histidine as a Proton Shuttle in Catalysis by Human Carbonic Anhydrase II
Biochemistry, 44, 2005
|
|
3UN3
 
 | | phosphopentomutase T85Q variant soaked with glucose 1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-alpha-D-glucopyranose, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Iverson, T.M, Birmingham, W.R, Panosian, T.D, Nannemann, D.P, Bachmann, B.O. | | Deposit date: | 2011-11-15 | | Release date: | 2012-02-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular Differences between a Mutase and a Phosphatase: Investigations of the Activation Step in Bacillus cereus Phosphopentomutase.
Biochemistry, 51, 2012
|
|
2I2H
 
 | | NMR structure of TPC3 in TFE | | Descriptor: | signaling peptide TCP3 | | Authors: | Syvitski, R.T, Jakeman, D.L, Li, Y. | | Deposit date: | 2006-08-16 | | Release date: | 2006-10-17 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | Structure-Activity Analysis of Quorum-Sensing Signaling Peptides from Streptococcus mutans.
J.Bacteriol., 189, 2007
|
|
1HQT
 
 | | THE CRYSTAL STRUCTURE OF AN ALDEHYDE REDUCTASE Y50F MUTANT-NADP COMPLEX AND ITS IMPLICATIONS FOR SUBSTRATE BINDING | | Descriptor: | ALDEHYDE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ye, Q, Hyndman, D, Green, N.C, Li, L, Korithoski, B, Jia, Z, Flynn, T.G. | | Deposit date: | 2000-12-19 | | Release date: | 2001-05-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of an Aldehyde Reductase Y50F Mutant-NADP Complex and its Implications for Substrate Binding
Chem.Biol.Interact., 132, 2001
|
|
1S0W
 
 | |
7TV9
 
 | | HUMAN COMPLEMENT COMPONENT C3B IN COMPLEX WITH APL-1030 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, APL-1030 Nanofitin, Complement C3 beta chain, ... | | Authors: | Fontano, E, Nadupalli, A, Lakshminarasimhan, D, White, A, Garlish, J, Cinier, M, Chevrel, A, Perrocheau, A, Eyerman, D, Orme, M, Kitten, O, Scheibler, L. | | Deposit date: | 2022-02-04 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Discovery of APL-1030, a Novel, High-Affinity Nanofitin Inhibitor of C3-Mediated Complement Activation.
Biomolecules, 12, 2022
|
|
7P6E
 
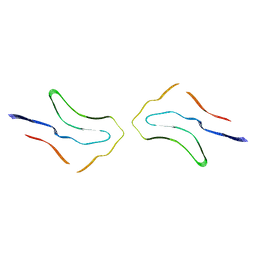 | | Argyrophilic grain disease type 2 tau filament | | Descriptor: | Microtubule-associated protein tau | | Authors: | Shi, Y, Zhang, W, Yang, Y, Murzin, A.G, Falcon, B, Kotecha, A, van Beers, M, Tarutani, A, Kametani, F, Garringer, H.J, Vidal, R, Hallinan, G.I, Lashley, T, Saito, Y, Murayama, S, Yoshida, M, Tanaka, H, Kakita, A, Ikeuchi, T, Robinson, A.C, Mann, D.M.A, Kovacs, G.G, Revesz, T, Ghetti, B, Hasegawa, M, Goedert, M, Scheres, S.H.W. | | Deposit date: | 2021-07-15 | | Release date: | 2021-09-15 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-based classification of tauopathies.
Nature, 598, 2021
|
|
2O0I
 
 | | crystal structure of the R185A mutant of the N-terminal domain of the Group B Streptococcus Alpha C protein | | Descriptor: | C protein alpha-antigen | | Authors: | Hogle, J.M, Filman, D.J, Baron, M.J, Madoff, L.C, Iglesias, A. | | Deposit date: | 2006-11-27 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Identification of a glycosaminoglycan binding region of the alpha C protein that mediates entry of group B streptococci into host cells.
J.Biol.Chem., 282, 2007
|
|
4FDX
 
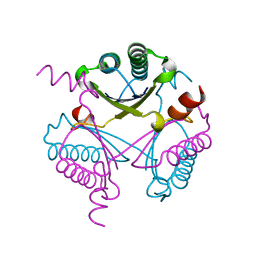 | |
3ZNV
 
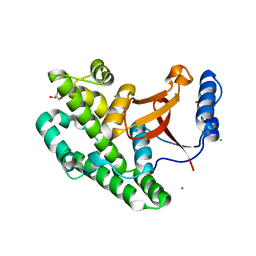 | | Crystal structure of the OTU domain of OTULIN at 1.3 Angstroms. | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Keusekotten, K, Elliott, P.R, Glockner, L, Kulathu, Y, Wauer, T, Krappmann, D, Hofmann, K, Komander, D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Otulin Antagonizes Lubac Signaling by Specifically Hydrolyzing met1-Linked Polyubiquitin.
Cell(Cambridge,Mass.), 153, 2013
|
|
3J48
 
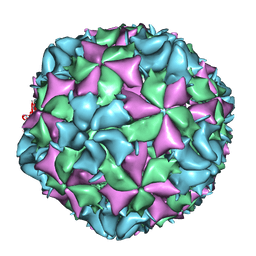 | | Cryo-EM structure of Poliovirus 135S particles | | Descriptor: | Protein VP1, Protein VP2, Protein VP3 | | Authors: | Butan, C, Fiman, D.J, Hogle, J.M. | | Deposit date: | 2013-06-28 | | Release date: | 2013-12-04 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Cryo-Electron Microscopy Reconstruction Shows Poliovirus 135S Particles Poised for Membrane Interaction and RNA Release.
J.Virol., 88, 2014
|
|
1NQ7
 
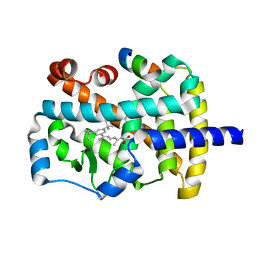 | | Characterization of ligands for the orphan nuclear receptor RORbeta | | Descriptor: | 7-(3,5-DITERT-BUTYLPHENYL)-3-METHYLOCTA-2,4,6-TRIENOIC ACID, NUCLEAR RECEPTOR ROR-BETA, STEROID RECEPTOR COACTIVATOR-1 | | Authors: | Stehlin-Gaon, C, Willmann, D, Sanglier, S, Van Dorsselaer, A, Renaud, J.-P, Moras, D, Schuele, R. | | Deposit date: | 2003-01-21 | | Release date: | 2003-09-23 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | All-trans retinoic acid is a ligand for the orphan nuclear receptor RORbeta
Nat.Struct.Biol., 10, 2003
|
|
1TEU
 
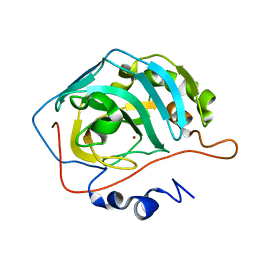 | | Effect of Shuttle Location and pH Environment on H+ Transfer in Human Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase II, ZINC ION | | Authors: | Fisher, Z, Hernandez Prada, J.A, Tu, C.K, Duda, D, Yoshioka, C, An, H, Govindasamy, L, Silverman, D.N, McKenna, R. | | Deposit date: | 2004-05-25 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Kinetic Characterization of Active-Site Histidine as a Proton Shuttle in Catalysis by Human Carbonic Anhydrase II
Biochemistry, 44, 2005
|
|
1TH1
 
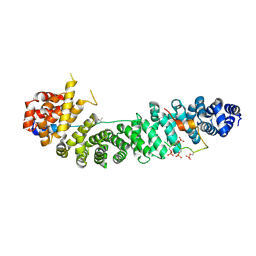 | | Beta-catenin in complex with a phosphorylated APC 20aa repeat fragment | | Descriptor: | Adenomatous polyposis coli protein, Beta-catenin | | Authors: | Xing, Y, Clements, W.K, Le Trong, I, Hinds, T.R, Stenkamp, R, Kimelman, D, Xu, W. | | Deposit date: | 2004-05-31 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a beta-Catenin/APC Complex Reveals a Critical Role for APC Phosphorylation in APC Function.
Mol.Cell, 15, 2004
|
|
4FAZ
 
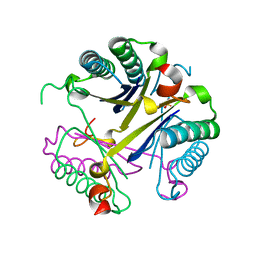 | |
3ZNX
 
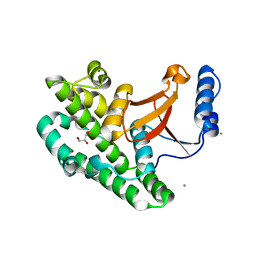 | | Crystal structure of the OTU domain of OTULIN D336A mutant | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Keusekotten, K, Elliott, P.R, Glockner, L, Kulathu, Y, Wauer, T, Krappmann, D, Hofmann, K, Komander, D. | | Deposit date: | 2013-02-18 | | Release date: | 2013-06-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Otulin Antagonizes Lubac Signaling by Specifically Hydrolyzing met1-Linked Polyubiquitin.
Cell(Cambridge,Mass.), 153, 2013
|
|
2WX1
 
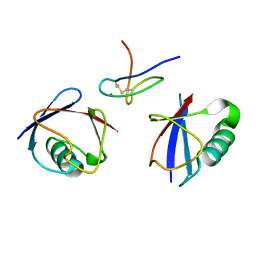 | | TAB2 NZF DOMAIN IN COMPLEX WITH Lys63-linked tri-ubiquitin, P212121 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 7-INTERACTING PROTEIN 2, UBIQUITIN, ZINC ION | | Authors: | Kulathu, Y, Akutsu, M, Bremm, A, Hofmann, K, Komander, D. | | Deposit date: | 2009-10-30 | | Release date: | 2009-11-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Two-Sided Ubiquitin Binding Explains Specificity of the Tab2 Nzf Domain
Nat.Struct.Mol.Biol., 16, 2009
|
|
1MOO
 
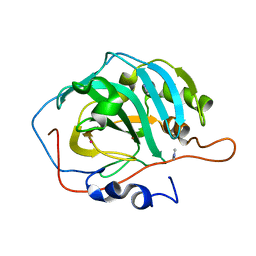 | | Site Specific Mutant (H64A) of Human Carbonic Anhydrase II at high resolution | | Descriptor: | 4-METHYLIMIDAZOLE, Carbonic Anhydrase II, MERCURY (II) ION, ... | | Authors: | Duda, D.M, Govindasamy, L, Agbandje-McKenna, M, Tu, C.K, Silverman, D.N, McKenna, R. | | Deposit date: | 2002-09-09 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | The refined atomic structure of carbonic anhydrase II at 1.05 A resolution: implications of chemical rescue of proton transfer.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1IX2
 
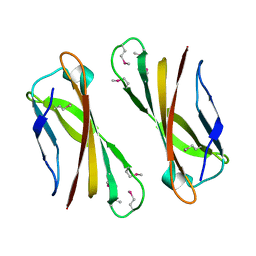 | | Crystal Structure of Selenomethionine PcoC, a Copper Resistance Protein from Escherichia coli | | Descriptor: | PcoC copper resistance protein | | Authors: | Wernimont, A.K, Huffman, D.L, Finney, L.A, Demeler, B, O'Halloran, T.V, Rosenzweig, A.C. | | Deposit date: | 2002-06-10 | | Release date: | 2002-11-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structure and dimerization equilibria of PcoC, a methionine-rich copper resistance protein from Escherichia coli
J.BIOL.INORG.CHEM., 8, 2003
|
|
