7N64
 
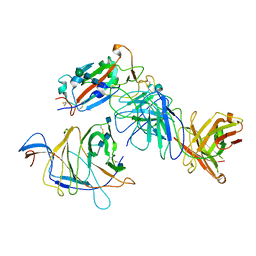 | | SARS-CoV-2 Spike (2P) in complex with G32R7 Fab (RBD and NTD local reconstruction) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, G32R7 Fab heavy chain, ... | | Authors: | Windsor, I.W, Jenni, S, Tong, P, Gautam, A.K, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-06-07 | | Release date: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Memory B cell repertoire for recognition of evolving SARS-CoV-2 spike.
Biorxiv, 2021
|
|
6EQI
 
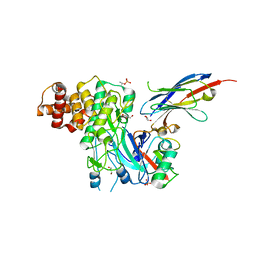 | | Structure of PINK1 bound to ubiquitin | | Descriptor: | GLYCEROL, Nb696, Serine/threonine-protein kinase PINK1, ... | | Authors: | Schubert, A.F, Gladkova, C, Pardon, E, Wagstaff, J.L, Freund, S.M.V, Steyaert, J, Maslen, S, Komander, D. | | Deposit date: | 2017-10-13 | | Release date: | 2017-11-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of PINK1 in complex with its substrate ubiquitin.
Nature, 552, 2017
|
|
8THV
 
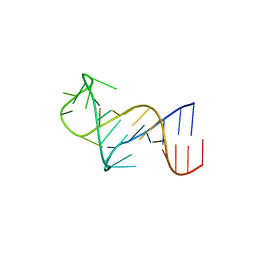 | | FARFAR-NMR ensemble of HIV-1 TAR with apical loop capturing ground and excited conformational states | | Descriptor: | RNA (29-MER) | | Authors: | Roy, R, Geng, A, Shi, H, Merriman, D.K, Dethoff, E.A, Salmon, L, Al-Hashimi, H.M. | | Deposit date: | 2023-07-18 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Kinetic Resolution of the Atomic 3D Structures Formed by Ground and Excited Conformational States in an RNA Dynamic Ensemble.
J.Am.Chem.Soc., 145, 2023
|
|
5TAR
 
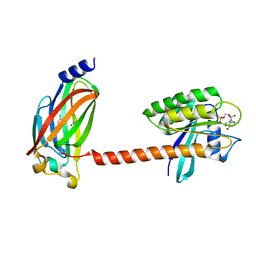 | | Crystal structure of farnesylated and methylated kras4b in complex with PDE-delta (crystal form II - with ordered hypervariable region) | | Descriptor: | 1,2-ETHANEDIOL, FARNESYL, GTPase KRas, ... | | Authors: | Dharmaiah, S, Tran, T.H, Simanshu, D.K. | | Deposit date: | 2016-09-10 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of recognition of farnesylated and methylated KRAS4b by PDE delta.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5RL4
 
 | | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors -- Crystal Structure of SARS-CoV-2 main protease in complex with LON-WEI-adc59df6-3 (Mpro-x3124) | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-(4-tert-butylphenyl)-N-[(1R)-2-(methylamino)-2-oxo-1-(pyridin-3-yl)ethyl]propanamide | | Authors: | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Zaidman, D, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Gorrie-Stone, T.J, Skyner, R, London, N, Walsh, M.A, von Delft, F. | | Deposit date: | 2020-08-05 | | Release date: | 2020-12-02 | | Last modified: | 2021-07-07 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | PanDDA analysis group deposition of computational designs of SARS-CoV-2 main protease covalent inhibitors
To Be Published
|
|
5TB5
 
 | | Crystal structure of full-length farnesylated and methylated KRAS4b in complex with PDE-delta (crystal form I - with partially disordered hypervariable region) | | Descriptor: | 1,2-ETHANEDIOL, FARNESYL, GTPase KRas, ... | | Authors: | Dharmaiah, S, Tran, T.H, Simanshu, D.K. | | Deposit date: | 2016-09-11 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of recognition of farnesylated and methylated KRAS4b by PDE delta.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5UI2
 
 | | CRYSTAL STRUCTURE OF ORANGE CAROTENOID PROTEIN | | Descriptor: | (3'R)-3'-hydroxy-beta,beta-caroten-4-one, CHLORIDE ION, Orange carotenoid-binding protein, ... | | Authors: | KERFELD, C.A, SAWAYA, M.R, VISHNU, B, KROGMANN, D, YEATES, T.O. | | Deposit date: | 2017-01-12 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a cyanobacterial water-soluble carotenoid binding protein.
Structure, 11, 2003
|
|
3X0W
 
 | | Crystal structure of PLEKHM1 LIR-fused human LC3B_2-119 | | Descriptor: | Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Suzuki, H, McEwan, D.G, Popovic, D, Gubas, A, Terawaki, S, Stadel, D, Coxon, F, Stegmann, D.M, Bhogaraju, S, Maddi, K, Kirchhoff, A, Gatti, E, Helfrich, M.H, Behrends, C, Pierre, P, Dikic, I, Wakatsuki, S. | | Deposit date: | 2014-10-22 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins.
Mol.Cell, 57, 2015
|
|
6CAA
 
 | | CryoEM structure of human SLC4A4 sodium-coupled acid-base transporter NBCe1 | | Descriptor: | Electrogenic sodium bicarbonate cotransporter 1 | | Authors: | Huynh, K.W, Jiang, J, Abuladze, N, Tsirulnikov, K, Kao, L, Shao, X, Newman, D, Azimov, R, Pushkin, A, Zhou, Z.H, Kurtz, I. | | Deposit date: | 2018-01-29 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | CryoEM structure of the human SLC4A4 sodium-coupled acid-base transporter NBCe1.
Nat Commun, 9, 2018
|
|
3ZJE
 
 | | A20 OTU domain in reversibly oxidised (SOH) state | | Descriptor: | 1,2-ETHANEDIOL, A20P50, CHLORIDE ION | | Authors: | Kulathu, Y, Garcia, F.J, Mevissen, T.E.T, Busch, M, Arnaudo, N, Carroll, K.S, Barford, D, Komander, D. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Regulation of A20 and Other Otu Deubiquitinases by Reversible Oxidation
Nat.Commun., 4, 2013
|
|
7LC2
 
 | |
7LC1
 
 | |
4C0J
 
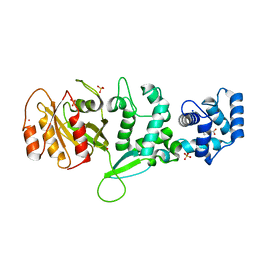 | | Crystal structure of Drosophila Miro EF hand and cGTPase domains in the apo state (Apo-MiroS) | | Descriptor: | L-HOMOSERINE, MITOCHONDRIAL RHO GTPASE, SODIUM ION, ... | | Authors: | Klosowiak, J.L, Focia, P.J, Wawrzak, Z, Chakravarthy, S, Landahl, E.C, Freymann, D.M, Rice, S.E. | | Deposit date: | 2013-08-05 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural Coupling of the EF Hand and C-Terminal Gtpase Domains in the Mitochondrial Protein Miro.
Embo Rep., 14, 2013
|
|
4C0K
 
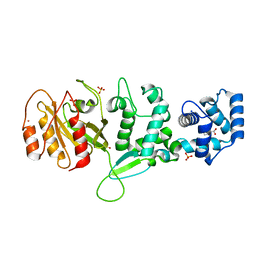 | | Crystal structure of Drosophila Miro EF hand and cGTPase domains bound to one calcium ion (Ca-MiroS) | | Descriptor: | CALCIUM ION, L-HOMOSERINE, MITOCHONDRIAL RHO GTPASE, ... | | Authors: | Klosowiak, J.L, Focia, P.J, Wawrzak, Z, Chakravarthy, S, Landahl, E.C, Freymann, D.M, Rice, S.E. | | Deposit date: | 2013-08-05 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Structural Coupling of the EF Hand and C-Terminal Gtpase Domains in the Mitochondrial Protein Miro.
Embo Rep., 14, 2013
|
|
4C0L
 
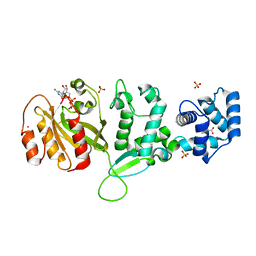 | | Crystal structure of Drosophila Miro EF hand and cGTPase domains bound to one magnesium ion and Mg:GDP (MgGDP-MiroS) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, L-HOMOSERINE, MAGNESIUM ION, ... | | Authors: | Klosowiak, J.L, Focia, P.J, Wawrzak, Z, Chakravarthy, S, Landahl, E.C, Freymann, D.M, Rice, S.E. | | Deposit date: | 2013-08-05 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Coupling of the EF Hand and C-Terminal Gtpase Domains in the Mitochondrial Protein Miro.
Embo Rep., 14, 2013
|
|
4BOS
 
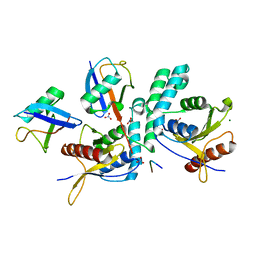 | | Structure of OTUD2 OTU domain in complex with Ubiquitin K11-linked peptide | | Descriptor: | MAGNESIUM ION, NITRATE ION, OTUD2, ... | | Authors: | Mevissen, T.E.T, Hospenthal, M.K, Geurink, P.P, Elliott, P.R, Akutsu, M, Arnaudo, N, Ekkebus, R, Kulathu, Y, Wauer, T, El Oualid, F, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2013-05-22 | | Release date: | 2013-07-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Otu Deubiquitinases Reveal Mechanisms of Linkage Specificity and Enable Ubiquitin Chain Restriction Analysis.
Cell(Cambridge,Mass.), 154, 2013
|
|
4BOZ
 
 | | Structure of OTUD2 OTU domain in complex with K11-linked di ubiquitin | | Descriptor: | GLYCEROL, UBIQUITIN THIOESTERASE OTU1, UBIQUITIN-C | | Authors: | Mevissen, T.E.T, Hospenthal, M.K, Geurink, P.P, Elliott, P.R, Akutsu, M, Arnaudo, N, Ekkebus, R, Kulathu, Y, Wauer, T, El Oualid, F, Freund, S.M.V, Ovaa, H, Komander, D. | | Deposit date: | 2013-05-22 | | Release date: | 2013-07-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Otu Deubiquitinases Reveal Mechanisms of Linkage Specificity and Enable Ubiquitin Chain Restriction Analysis.
Cell(Cambridge,Mass.), 154, 2013
|
|
3ZNH
 
 | | Crimean Congo Hemorrhagic Fever Virus OTU domain in complex with ubiquitin-propargyl. | | Descriptor: | POLYUBIQUITIN-B, UBIQUITIN THIOESTERASE | | Authors: | Ekkebus, R, vanKasteren, S.I, Kulathu, Y, Scholten, A, Berlin, I, deJong, A, Goerdayal, G, Neefjes, J, Heck, A.J.R, Komander, D, Ovaa, H. | | Deposit date: | 2013-02-14 | | Release date: | 2013-02-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | On Terminal Alkynes that Can React with Active-Site Cysteine Nucleophiles in Proteases.
J.Am.Chem.Soc., 135, 2013
|
|
3ZJD
 
 | | A20 OTU domain in reduced, active state at 1.87 A resolution | | Descriptor: | 1,2-ETHANEDIOL, A20P50, CHLORIDE ION | | Authors: | Kulathu, Y, Garcia, F.J, Mevissen, T.E.T, Busch, M, Arnaudo, N, Carroll, K.S, Barford, D, Komander, D. | | Deposit date: | 2013-01-17 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Regulation of A20 and Other Otu Deubiquitinases by Reversible Oxidation
Nat.Commun., 4, 2013
|
|
7MW1
 
 | | Crystal structure of the Homo sapiens NUP93-NUP53 complex (NUP93 residues 174-819; NUP53 residues 84-150) | | Descriptor: | Nuclear pore complex protein Nup93, Nucleoporin Nup35 | | Authors: | Petrovic, S, Samanta, D, Perriches, T, Bley, C.J, Thierbach, K, Brown, B, Nie, S, Mobbs, G.W, Stevens, T.A, Liu, X, Tomaleri, G.P, Schaus, L, Hoelz, A. | | Deposit date: | 2021-05-15 | | Release date: | 2022-06-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Architecture of the linker-scaffold in the nuclear pore.
Science, 376, 2022
|
|
3ZRH
 
 | | Crystal structure of the Lys29, Lys33-linkage-specific TRABID OTU deubiquitinase domain reveals an Ankyrin-repeat ubiquitin binding domain (AnkUBD) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, UBIQUITIN THIOESTERASE ZRANB1 | | Authors: | Licchesi, J.D.F, Akutsu, M, Komander, D. | | Deposit date: | 2011-06-16 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | An Ankyrin-Repeat Ubiquitin-Binding Domain Determines Trabid'S Specificity for Atypical Ubiquitin Chains.
Nat.Struct.Mol.Biol., 19, 2011
|
|
4AE3
 
 | | Crystal structure of ammosamide 272:myosin-2 motor domain complex | | Descriptor: | 1,2-ETHANEDIOL, ADP ORTHOVANADATE, AMMOSAMIDE 272, ... | | Authors: | Chinthalapudi, K, Heissler, S.M, Fenical, W, Manstein, D.J. | | Deposit date: | 2012-01-05 | | Release date: | 2013-01-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Ammosamide Mediated Myosin Motor Activity Inhibition
To be Published
|
|
433D
 
 | |
4BM9
 
 | |
7N62
 
 | | SARS-CoV-2 Spike (2P) in complex with C12C9 Fab (NTD local reconstruction) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C12C9 Fab heavy chain, C12C9 Fab light chain, ... | | Authors: | Windsor, I.W, Jenni, S, Bajic, G, Tong, P, Gautam, A.K, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-06-07 | | Release date: | 2021-08-04 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Memory B cell repertoire for recognition of evolving SARS-CoV-2 spike.
Biorxiv, 2021
|
|
