4RHB
 
 | |
4RH8
 
 | |
7VDQ
 
 | | The structure of cyclin-dependent kinase 5 (CDK5) in complex with p25 and Compound 7 | | Descriptor: | 2-[[7-[[2-fluoranyl-4-[3-(hydroxymethyl)pyrazol-1-yl]phenyl]amino]-1,6-naphthyridin-2-yl]-(1-methylpiperidin-4-yl)amino]ethanoic acid, Cyclin-dependent kinase 5 activator 1, p25, ... | | Authors: | Malojcic, G, Clugston, S.L, Daniels, M, Harmange, J.C, Ledeborer, M. | | Deposit date: | 2021-09-07 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Discovery and Optimization of Highly Selective Inhibitors of CDK5.
J.Med.Chem., 65, 2022
|
|
7VDP
 
 | | The structure of cyclin-dependent kinase 5 (CDK5) in complex with p25 and Compound 1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cyclin-dependent kinase 5 activator 1, ... | | Authors: | Malojcic, G, Clugston, S.L, Daniels, M, Harmange, J.C, Ledeborer, M. | | Deposit date: | 2021-09-07 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Discovery and Optimization of Highly Selective Inhibitors of CDK5.
J.Med.Chem., 65, 2022
|
|
7VDS
 
 | | The structure of cyclin-dependent kinase 5 (CDK5) in complex with p25 and Compound 24 | | Descriptor: | 1,2-ETHANEDIOL, 5-fluoranyl-4-[[2-[(1R)-1-(1-methylpiperidin-4-yl)-1-oxidanyl-ethyl]-1,6-naphthyridin-7-yl]amino]-2-morpholin-4-yl-benzenecarbonitrile, CHLORIDE ION, ... | | Authors: | Malojcic, G, Clugston, S.L, Daniels, M, Harmange, J.C, Ledeborer, M. | | Deposit date: | 2021-09-07 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Discovery and Optimization of Highly Selective Inhibitors of CDK5.
J.Med.Chem., 65, 2022
|
|
7VDR
 
 | | The structure of cyclin-dependent kinase 5 (CDK5) in complex with p25 and Compound 13 | | Descriptor: | (1R)-1-[7-[(2-fluoranyl-4-pyrazol-1-yl-phenyl)amino]-1,6-naphthyridin-2-yl]-1-(1-methylpiperidin-4-yl)ethanol, 1,2-ETHANEDIOL, Cyclin-dependent kinase 5 activator 1, ... | | Authors: | Malojcic, G, Clugston, S.L, Daniels, M, Harmange, J.C, Ledeborer, M. | | Deposit date: | 2021-09-07 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery and Optimization of Highly Selective Inhibitors of CDK5.
J.Med.Chem., 65, 2022
|
|
3ELQ
 
 | | Crystal structure of a bacterial arylsulfate sulfotransferase | | Descriptor: | Arylsulfate sulfotransferase, CHLORIDE ION, SULFATE ION | | Authors: | Malojcic, G, Owen, R.L, Grimshaw, J.P, Glockshuber, R. | | Deposit date: | 2008-09-23 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural and biochemical basis for PAPS-independent sulfuryl transfer by aryl sulfotransferase from uropathogenic Escherichia coli.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3ETT
 
 | | Crystal structure of a bacterial arylsulfate sulfotransferase catalytic intermediate with 4-nitrophenol bound in the active site | | Descriptor: | Arylsulfate sulfotransferase, P-NITROPHENOL, SULFATE ION | | Authors: | Malojcic, G, Owen, R.L, Grimshaw, J.P, Glockshuber, R. | | Deposit date: | 2008-10-08 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural and biochemical basis for PAPS-independent sulfuryl transfer by aryl sulfotransferase from uropathogenic Escherichia coli.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
3ETS
 
 | | Crystal structure of a bacterial arylsulfate sulfotransferase catalytic intermediate with 4-methylumbelliferone bound in the active site | | Descriptor: | 7-hydroxy-4-methyl-2H-chromen-2-one, Arylsulfate sulfotransferase, SULFATE ION | | Authors: | Malojcic, G, Owen, R.L, Grimshaw, J.P, Glockshuber, R. | | Deposit date: | 2008-10-08 | | Release date: | 2008-11-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A structural and biochemical basis for PAPS-independent sulfuryl transfer by aryl sulfotransferase from uropathogenic Escherichia coli.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
4P04
 
 | |
4P06
 
 | |
7VDU
 
 | | The structure of cyclin-dependent kinase 2 (CDK2) in complex with Compound 1 | | Descriptor: | Cyclin-dependent kinase 2, [1-[3-fluoranyl-4-[(2-piperidin-4-yloxy-1,6-naphthyridin-7-yl)amino]phenyl]pyrazol-3-yl]methanol | | Authors: | Malojcic, G, Clugston, S.L, Daniels, M, Harmange, J.C, Ledeborer, M. | | Deposit date: | 2021-09-07 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Discovery and Optimization of Highly Selective Inhibitors of CDK5.
J.Med.Chem., 65, 2022
|
|
4P05
 
 | |
3E9J
 
 | |
4P07
 
 | |
4NHR
 
 | |
7RA5
 
 | | CDK2 IN COMPLEX WITH COMPOUND 4 | | Descriptor: | 4-[7-(methanesulfonyl)-1H-indol-3-yl]-N-[(3S)-piperidin-3-yl]-5-(trifluoromethyl)pyrimidin-2-amine, Cyclin-dependent kinase 2 | | Authors: | Marineau, J.J, Malojcic, G. | | Deposit date: | 2021-06-30 | | Release date: | 2021-11-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery of SY-5609: A Selective, Noncovalent Inhibitor of CDK7.
J.Med.Chem., 65, 2022
|
|
4M8G
 
 | | Crystal structure of Se-Met hN33/Tusc3 | | Descriptor: | Tumor suppressor candidate 3 | | Authors: | Mohorko, E, Owen, R.L, Malojcic, G, Brozzo, M.S, Aebi, M, Glockshuber, R. | | Deposit date: | 2013-08-13 | | Release date: | 2014-03-26 | | Last modified: | 2014-05-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of substrate specificity of human oligosaccharyl transferase subunit n33/tusc3 and its role in regulating protein N-glycosylation.
Structure, 22, 2014
|
|
4M90
 
 | | crystal structure of oxidized hN33/Tusc3 | | Descriptor: | Tumor suppressor candidate 3 | | Authors: | Mohorko, E, Owen, R.L, Malojcic, G, Brozzo, M.S, Aebi, M, Glockshuber, R. | | Deposit date: | 2013-08-14 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of substrate specificity of human oligosaccharyl transferase subunit n33/tusc3 and its role in regulating protein N-glycosylation.
Structure, 22, 2014
|
|
4M92
 
 | | Crystal structure of hN33/Tusc3-peptide 2 | | Descriptor: | Interleukin-1 receptor accessory protein-like 1, Tumor suppressor candidate 3 | | Authors: | Mohorko, E, Owen, R.L, Malojcic, G, Brozzo, M.S, Aebi, M, Glockshuber, R. | | Deposit date: | 2013-08-14 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of substrate specificity of human oligosaccharyl transferase subunit n33/tusc3 and its role in regulating protein N-glycosylation.
Structure, 22, 2014
|
|
4M91
 
 | | crystal structure of hN33/Tusc3-peptide 1 | | Descriptor: | Protein cereblon, Tumor suppressor candidate 3 | | Authors: | Mohorko, E, Owen, R.L, Malojcic, G, Brozzo, M.S, Aebi, M, Glockshuber, R. | | Deposit date: | 2013-08-14 | | Release date: | 2014-03-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis of substrate specificity of human oligosaccharyl transferase subunit n33/tusc3 and its role in regulating protein N-glycosylation.
Structure, 22, 2014
|
|
7B16
 
 | | TRPC4 in complex with inhibitor GFB-9289 | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, 5-chloranyl-4-(4-cyclohexyl-3-oxidanylidene-piperazin-1-yl)-1~{H}-pyridazin-6-one, CALCIUM ION, ... | | Authors: | Vinayagam, D, Quentin, D, Sistel, O, Merino, F, Stabrin, M, Hofnagel, O, Ledeboer, M.W, Malojcic, G, Raunser, S. | | Deposit date: | 2020-11-23 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis of TRPC4 regulation by calmodulin and pharmacological agents.
Elife, 9, 2020
|
|
7B0S
 
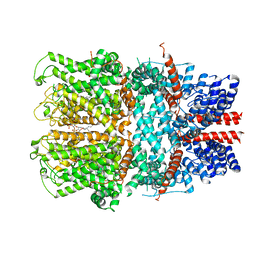 | | TRPC4 in complex with inhibitor GFB-8438 | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, 5-chloranyl-4-[3-oxidanylidene-4-[[2-(trifluoromethyl)phenyl]methyl]piperazin-1-yl]-1~{H}-pyridazin-6-one, CALCIUM ION, ... | | Authors: | Vinayagam, D, Quentin, D, Sistel, O, Merino, F, Stabrin, M, Hofnagel, O, Ledeboer, M.W, Malojcic, G, Raunser, S. | | Deposit date: | 2020-11-21 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of TRPC4 regulation by calmodulin and pharmacological agents.
Elife, 9, 2020
|
|
7B1G
 
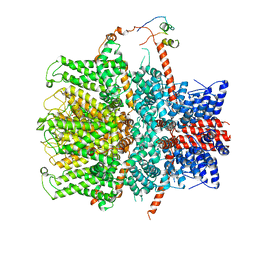 | | TRPC4 in complex with Calmodulin | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, CALCIUM ION, Calmodulin-1, ... | | Authors: | Vinayagam, D, Quentin, D, Sistel, O, Merino, F, Stabrin, M, Hofnagel, O, Ledeboer, M.W, Malojcic, G, Raunser, S. | | Deposit date: | 2020-11-24 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of TRPC4 regulation by calmodulin and pharmacological agents.
Elife, 9, 2020
|
|
7B0J
 
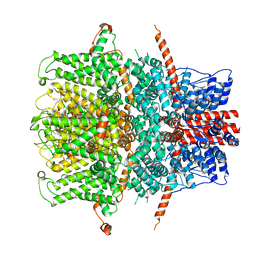 | | TRPC4 in LMNG detergent | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, CALCIUM ION, Transient receptor potential cation channel subfamily c member 4a | | Authors: | Vinayagam, D, Quentin, D, Sistel, O, Merino, F, Stabrin, M, Hofnagel, O, Ledeboer, M.W, Malojcic, G, Raunser, S. | | Deposit date: | 2020-11-20 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of TRPC4 regulation by calmodulin and pharmacological agents.
Elife, 9, 2020
|
|
