2J9K
 
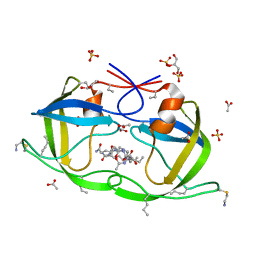 | | Atomic-resolution Crystal Structure of Chemically-Synthesized HIV-1 Protease Complexed with Inhibitor MVT-101 | | 分子名称: | ACETATE ION, GLYCEROL, N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, ... | | 著者 | Malito, E, Shen, Y, Johnson, E.C.B, Tang, W.J. | | 登録日 | 2006-11-11 | | 公開日 | 2007-08-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Insights from Atomic-Resolution X-Ray Structures of Chemically Synthesized HIV-1 Protease in Complex with Inhibitors.
J.Mol.Biol., 373, 2007
|
|
2JG4
 
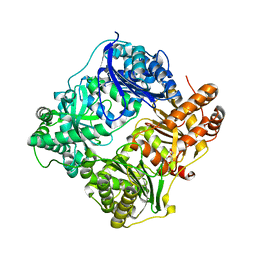 | | Substrate-free IDE structure in its closed conformation | | 分子名称: | 1,4-DIETHYLENE DIOXIDE, INSULIN DEGRADING ENZYME, ZINC ION | | 著者 | Malito, E, Tang, W.J. | | 登録日 | 2007-02-07 | | 公開日 | 2007-07-03 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structure of Substrate-Free Human Insulin Degrading Enzyme (Ide) and Biophysical Analysis of ATP-Induced Conformational Switch of Ide
J.Biol.Chem., 282, 2007
|
|
2JE4
 
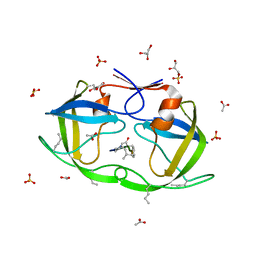 | | Atomic-resolution crystal structure of chemically-synthesized HIV-1 protease in complex with JG-365 | | 分子名称: | ACETATE ION, GLYCEROL, INHIBITOR MOLECULE JG365, ... | | 著者 | Malito, E, Johnson, E.C.B, Tang, W.J. | | 登録日 | 2007-01-15 | | 公開日 | 2007-08-28 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.07 Å) | | 主引用文献 | Modular Total Chemical Synthesis of a Human Immunodeficiency Virus Type 1 Protease.
J.Am.Chem.Soc., 129, 2007
|
|
4AE0
 
 | |
4BIG
 
 | | Crystal structure of the conserved staphylococcal antigen 1B, Csa1B | | 分子名称: | UNCHARACTERIZED LIPOPROTEIN SAOUHSC_00053 | | 著者 | Malito, E, Bottomley, M.J, Schluepen, C, Liberatori, S. | | 登録日 | 2013-04-10 | | 公開日 | 2013-08-07 | | 最終更新日 | 2013-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.274 Å) | | 主引用文献 | Mining the Bacterial Unknown Proteome: Identification and Characterization of a Novel Family of Highly Conserved Protective Antigens in Staphylococcus Aureus
Biochem.J., 455, 2013
|
|
4CJD
 
 | | Crystal structure of Neisseria meningitidis trimeric autotransporter and vaccine antigen NadA | | 分子名称: | IODIDE ION, NADA | | 著者 | Malito, E, Biancucci, M, Spraggon, G, Bottomley, M.J. | | 登録日 | 2013-12-19 | | 公開日 | 2014-11-26 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.056 Å) | | 主引用文献 | Structure of the Meningococcal Vaccine Antigen Nada and Epitope Mapping of a Bactericidal Antibody.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4BIH
 
 | | Crystal structure of the conserved staphylococcal antigen 1A, Csa1A | | 分子名称: | CALCIUM ION, UNCHARACTERIZED LIPOPROTEIN SAOUHSC_00053 | | 著者 | Malito, E, Bottomley, M.J, Spraggon, G, Schluepen, C, Liberatori, S. | | 登録日 | 2013-04-10 | | 公開日 | 2013-08-07 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.459 Å) | | 主引用文献 | Mining the Bacterial Unknown Proteome: Identification and Characterization of a Novel Family of Highly Conserved Protective Antigens in Staphylococcus Aureus
Biochem.J., 455, 2013
|
|
4B8Y
 
 | | Ferrichrome-bound FhuD2 | | 分子名称: | 1,2-ETHANEDIOL, CHLORIDE ION, FE (III) ION, ... | | 著者 | Malito, E, Bottomley, M.J, Spraggon, G. | | 登録日 | 2012-08-31 | | 公開日 | 2012-11-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural and Functional Characterization of the Staphylococcus Aureus Virulence Factor and Vaccine Candidate Fhud2.
Biochem.J., 449, 2013
|
|
4D7W
 
 | | Crystal structure of sortase C1 (SrtC1) from Streptococcus agalactiae | | 分子名称: | 1,2-ETHANEDIOL, SORTASE FAMILY PROTEIN | | 著者 | Malito, E, Lazzarin, M, Cozzi, R, Bottomley, M.J. | | 登録日 | 2014-11-28 | | 公開日 | 2015-08-19 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Noncanonical Sortase-Mediated Assembly of Pilus Type 2B in Group B Streptococcus.
Faseb J., 29, 2015
|
|
4DMW
 
 | | Crystal structure of the GT domain of Clostridium difficile toxin A (TcdA) in complex with UDP and Manganese | | 分子名称: | MANGANESE (II) ION, Toxin A, URIDINE-5'-DIPHOSPHATE | | 著者 | Malito, E, D'Urzo, N, Bottomley, M.J, Biancucci, M, Scarselli, M, Maione, D, Martinelli, M. | | 登録日 | 2012-02-08 | | 公開日 | 2012-07-18 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The structure of Clostridium difficile toxin A glucosyltransferase domain bound to Mn2+ and UDP provides insights into glucosyltransferase activity and product release.
Febs J., 279, 2012
|
|
4DMV
 
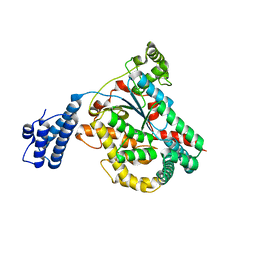 | | Crystal structure of the GT domain of Clostridium difficile Toxin A | | 分子名称: | Toxin A | | 著者 | Malito, E, D'Urzo, N, Bottomley, M.J, Biancucci, M, Maione, D, Scarselli, M, Martinelli, M. | | 登録日 | 2012-02-08 | | 公開日 | 2012-07-18 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | The structure of Clostridium difficile toxin A glucosyltransferase domain bound to Mn2+ and UDP provides insights into glucosyltransferase activity and product release.
Febs J., 279, 2012
|
|
7MPG
 
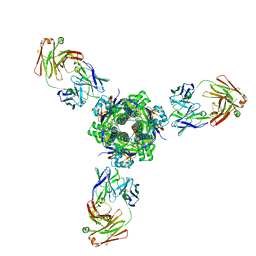 | |
7MMN
 
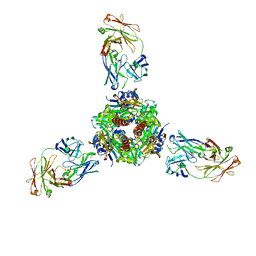 | |
7NRU
 
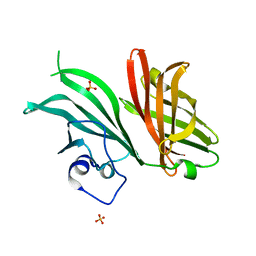 | |
7SBZ
 
 | |
7SA6
 
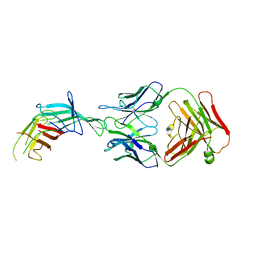 | | fHbp mutant 2416 bound to Fab JAR5 | | 分子名称: | Factor H-binding protein 2416, JAR5 Heavy Chain, JAR5 Light Chain | | 著者 | Chesterman, C, Malito, E, Bottomley, M.J. | | 登録日 | 2021-09-22 | | 公開日 | 2022-10-05 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Active Learning for Rapid Design: An iterative AI approach for accelerated vaccine design that combines active machine learning and high-throughput experimental evaluation
To Be Published
|
|
5AIP
 
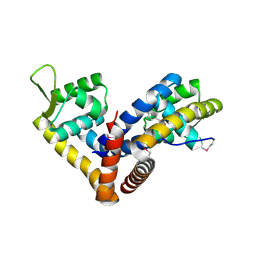 | | Crystal structure of NadR in complex with 4-hydroxyphenylacetate | | 分子名称: | 4-HYDROXYPHENYLACETATE, TRANSCRIPTIONAL REGULATOR, MARR FAMILY | | 著者 | Liguori, A, Malito, E, Bottomley, M.J. | | 登録日 | 2015-02-16 | | 公開日 | 2016-03-02 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Molecular Basis of Ligand-Dependent Regulation of Nadr, the Transcriptional Repressor of Meningococcal Virulence Factor Nada.
Plos Pathog., 12, 2016
|
|
5AIQ
 
 | |
4OXR
 
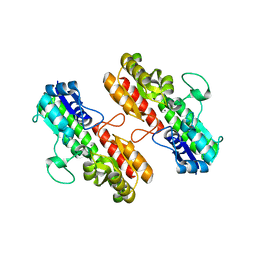 | | Structure of Staphylococcus pseudintermedius metal-binding protein SitA in complex with Manganese | | 分子名称: | MANGANESE (II) ION, Manganese ABC transporter, periplasmic-binding protein SitA | | 著者 | Abate, F, Malito, E, Bottomley, M. | | 登録日 | 2014-02-06 | | 公開日 | 2014-10-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Apo, Zn2+-bound and Mn2+-bound structures reveal ligand-binding properties of SitA from the pathogen Staphylococcus pseudintermedius.
Biosci.Rep., 34, 2014
|
|
4OXQ
 
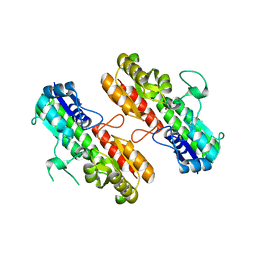 | | Structure of Staphylococcus pseudintermedius metal-binding protein SitA in complex with Zinc | | 分子名称: | Manganese ABC transporter, periplasmic-binding protein SitA, ZINC ION | | 著者 | Abate, F, Malito, E, Bottomley, M. | | 登録日 | 2014-02-06 | | 公開日 | 2014-10-29 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.62 Å) | | 主引用文献 | Apo, Zn2+-bound and Mn2+-bound structures reveal ligand-binding properties of SitA from the pathogen Staphylococcus pseudintermedius.
Biosci.Rep., 34, 2014
|
|
5M63
 
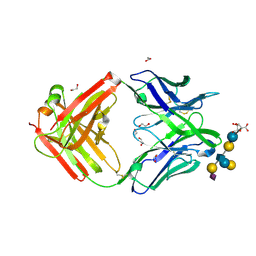 | | Crystal structure of group B Streptococcus type III DP2 oligosaccharide bound to Fab NVS-1-19-5 | | 分子名称: | (2~{R},3~{R},4~{S},5~{S})-2-[bis(oxidanyl)methyl]-5-(hydroxymethyl)oxolane-3,4-diol, 1,2-ETHANEDIOL, H chain of Fab NVS-1-19-5, ... | | 著者 | Carboni, F, Adamo, R, Veggi, D, Rappuoli, R, Malito, E, Margarit, I.R, Berti, F. | | 登録日 | 2016-10-24 | | 公開日 | 2017-05-03 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.74 Å) | | 主引用文献 | Structure of a protective epitope of group B Streptococcus type III capsular polysaccharide.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3CQF
 
 | |
5N4G
 
 | |
5N4J
 
 | |
6EUN
 
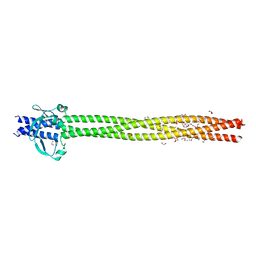 | | Crystal structure of Neisseria meningitidis vaccine antigen NadA variant 3 | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, ... | | 著者 | Dello Iacono, L, Liguori, A, Malito, E, Bottomley, M.J. | | 登録日 | 2017-10-30 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.45 Å) | | 主引用文献 | NadA3 Structures Reveal Undecad Coiled Coils and LOX1 Binding Regions Competed by Meningococcus B Vaccine-Elicited Human Antibodies.
MBio, 9, 2018
|
|
