8RQH
 
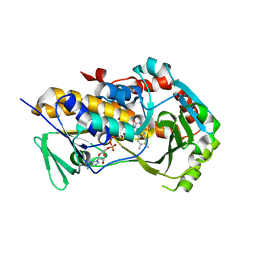 | | Crystal Structure of the flavoprotein monooxygenase TrlE from Streptomyces cyaneofuscatus Soc7 | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Sowa, S.T, Hoeing, L.S, Jakob, R.P, Maier, T, Teufel, R. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biosynthesis of the bacterial antibiotic 3,7-dihydroxytropolone through enzymatic salvaging of catabolic shunt products.
Chem Sci, 15, 2024
|
|
7R04
 
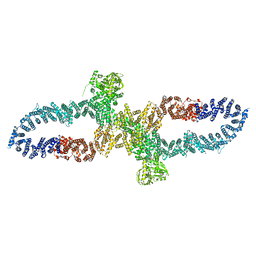 | | Neurofibromin in open conformation | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, Isoform I of Neurofibromin | | Authors: | Chaker-Margot, M, Scheffzek, K, Maier, T. | | Deposit date: | 2022-02-01 | | Release date: | 2022-03-30 | | Last modified: | 2022-04-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of activation of the tumor suppressor protein neurofibromin.
Mol.Cell, 82, 2022
|
|
7R03
 
 | |
3J9G
 
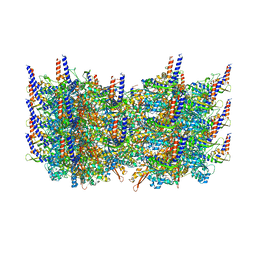 | | Atomic model of the VipA/VipB, the type six secretion system contractile sheath of Vibrio cholerae from cryo-EM | | Descriptor: | VipA, VipB | | Authors: | Kudryashev, M, Wang, R.Y.-R, Brackmann, M, Scherer, S, Maier, T, Baker, D, DiMaio, F, Stahlberg, H, Egelman, E.H, Basler, M. | | Deposit date: | 2015-01-16 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Type VI Secretion System Contractile Sheath.
Cell(Cambridge,Mass.), 160, 2015
|
|
3MF1
 
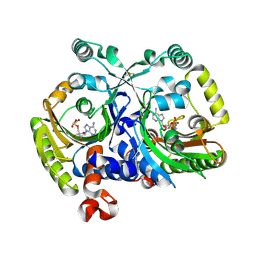 | | Crystal structure of class II aaRS homologue (Bll0957) complexed with an analogue of glycyl adenylate | | Descriptor: | 5'-O-(glycylsulfamoyl)adenosine, Bll0957 protein, ZINC ION | | Authors: | Weygand-Durasevic, I, Mocibob, M, Ivic, N, Bilokapic, S, Maier, T, Luic, M, Ban, N. | | Deposit date: | 2010-04-01 | | Release date: | 2010-07-28 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Homologs of aminoacyl-tRNA synthetases acylate carrier proteins and provide a link between ribosomal and nonribosomal peptide synthesis
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6QGY
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with nanobody B12 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, NanoB12, Outer membrane protein assembly factor BamA | | Authors: | Hartmann, J.-B, Kaur, H, Jakob, R.P, Zahn, M, Zimmermann, I, Seeger, M, Maier, T, Hiller, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach.
J.Biomol.Nmr, 73, 2019
|
|
5MUC
 
 | | Crystal structure of the FimH lectin domain in complex with 1,5-Anhydromannitol | | Descriptor: | 1-deoxy-alpha-D-mannopyranose, Protein FimH | | Authors: | Jakob, R.P, Rabbani, S, Ernst, B, Maier, T. | | Deposit date: | 2017-01-13 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | KinITC-One Method Supports both Thermodynamic and Kinetic SARs as Exemplified on FimH Antagonists.
Chemistry, 24, 2018
|
|
7R1V
 
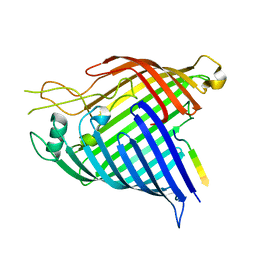 | |
7R1W
 
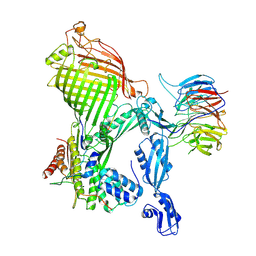 | | E. coli BAM complex (BamABCDE) bound to dynobactin A | | Descriptor: | Dynobactin A, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Jakob, R.P, Hiller, S, Maier, T. | | Deposit date: | 2022-02-03 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Computational identification of a systemic antibiotic for gram-negative bacteria.
Nat Microbiol, 7, 2022
|
|
7AYN
 
 | | Crystal structure of the lectin domain of the FimH variant Arg98Ala, in complex with Methyl 3-chloro-4-D-mannopyranosyloxy-3-biphenylcarboxylate | | Descriptor: | 1,2-ETHANEDIOL, Type 1 fimbrin D-mannose specific adhesin, methyl 3-[3-chloranyl-4-[(2~{R},3~{S},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-phenyl]benzoate | | Authors: | Jakob, R.P, Tomasic, T, Rabbani, S, Reisner, A, Jakopin, Z, Maier, T, Ernst, B, Anderluh, M. | | Deposit date: | 2020-11-12 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Does targeting Arg98 of FimH lead to high affinity antagonists?
Eur.J.Med.Chem., 211, 2020
|
|
4CST
 
 | | Crystal structure of FimH in complex with 3'-Chloro-4'-(alpha-D-mannopyranosyloxy)-biphenyl-4-carbonitrile | | Descriptor: | 3'-chloro-4'-(alpha-D-mannopyranosyloxy)biphenyl-4-carbonitrile, PROTEIN FIMH | | Authors: | Kleeb, S, Pang, L, Mayer, K, Sigl, A, Eris, D, Preston, R.C, Zihlmann, P, Abgottspon, D, Hutter, A, Scharenberg, M, Jian, X, Navarra, G, Rabbani, S, Smiesko, M, Luedin, N, Jakob, R.P, Schwardt, O, Maier, T, Sharpe, T, Ernst, B. | | Deposit date: | 2014-03-10 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Fimh Antagonists: Bioisosteres to Improve the in Vitro and in Vivo Pk/Pd Profile.
J.Med.Chem., 58, 2015
|
|
4CSS
 
 | | Crystal structure of FimH in complex with a sulfonamide biphenyl alpha D-mannoside | | Descriptor: | 4'-(alpha-D-Mannopyranosyloxy)-biphenyl-4-methyl sulfonamide, GLYCEROL, PROTEIN FIMH | | Authors: | Kleeb, S, Pang, L, Mayer, K, Sigl, A, Eris, D, Preston, R.C, Zihlmann, P, Abgottspon, D, Hutter, A, Scharenberg, M, Jian, X, Navarra, G, Rabbani, S, Smiesko, M, Luedin, N, Jakob, R.P, Schwardt, O, Maier, T, Sharpe, T, Ernst, B. | | Deposit date: | 2014-03-10 | | Release date: | 2015-02-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.069 Å) | | Cite: | Fimh Antagonists: Bioisosteres to Improve the in Vitro and in Vivo Pk/Pd Profile.
J.Med.Chem., 58, 2015
|
|
1CE7
 
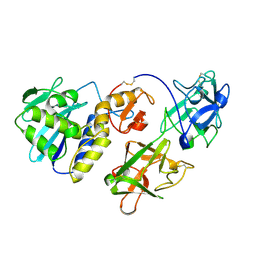 | | MISTLETOE LECTIN I FROM VISCUM ALBUM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (RIBOSOME-INACTIVATING PROTEIN TYPE II) | | Authors: | Krauspenhaar, R, Eschenburg, S, Perbandt, M, Kornilov, V, Konareva, N, Mikailova, I, Stoeva, S, Wacker, R, Maier, T, Singh, T.P, Mikhailov, A, Voelter, W, Betzel, C. | | Deposit date: | 1999-03-18 | | Release date: | 2000-03-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of mistletoe lectin I from Viscum album.
Biochem.Biophys.Res.Commun., 257, 1999
|
|
2Z9A
 
 | |
1OI8
 
 | | 5'-Nucleotidase (E. coli) with an Engineered Disulfide Bridge (P90C, L424C) | | Descriptor: | CARBONATE ION, MANGANESE (II) ION, PROTEIN USHA, ... | | Authors: | Schultz-Heienbrok, R, Maier, T, Straeter, N. | | Deposit date: | 2003-06-10 | | Release date: | 2004-06-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Trapping a 96 Degree Domain Rotation in Two Distinct Conformations by Engineered Disulfide Bridges
Protein Sci., 13, 2004
|
|
1OIE
 
 | |
4C16
 
 | | E-selectin lectin, EGF-like and two SCR domains complexed with glycomimetic antagonist | | Descriptor: | (1R,2R,3S)-3-methylcyclohexane-1,2-diol, (S)-CYCLOHEXYL LACTIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Preston, R.C, Jakob, R.P, Binder, F.P.C, Sager, C.P, Ernst, B, Maier, T. | | Deposit date: | 2013-08-09 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | E-Selectin Ligand Complexes Adopt an Extended High-Affinity Conformation.
J.Mol.Cell.Biol., 8, 2016
|
|
4BZA
 
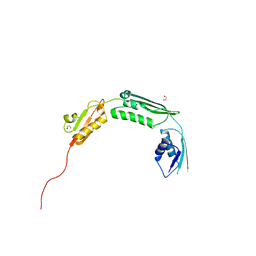 | | Crystal structure of TamA POTRA domains 1-3 from E. coli | | Descriptor: | 1,2-ETHANEDIOL, TRANSLOCATION AND ASSEMBLY MODULE TAMA | | Authors: | Jakob, R.P, Gruss, F, Zaehringer, F, Burmann, B.M, Hiller, S, Maier, T. | | Deposit date: | 2013-07-24 | | Release date: | 2013-09-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.839 Å) | | Cite: | The Structural Basis of Autotransporter Translocation by Tama
Nat.Struct.Mol.Biol., 20, 2013
|
|
1OID
 
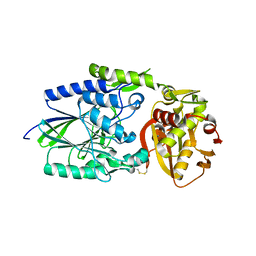 | |
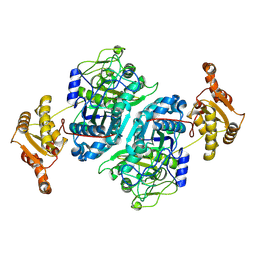
7ZMD
 
 | |
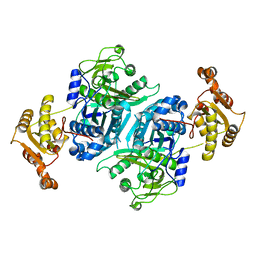
7ZMC
 
 | |
7ZMA
 
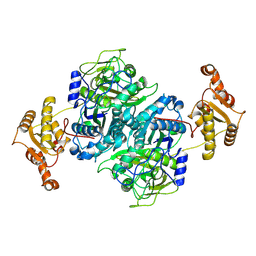 | |
7ZM9
 
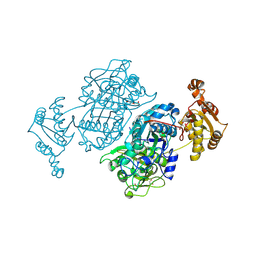 | | Ketosynthase domain 3 of Brevibacillus Brevis orphan BGC11 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Putative polyketide synthase | | Authors: | Tittes, Y.U, Herbst, D.A, Jakob, R.P, Maier, T. | | Deposit date: | 2022-04-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | The structure of a polyketide synthase bimodule core.
Sci Adv, 8, 2022
|
|
7ZMF
 
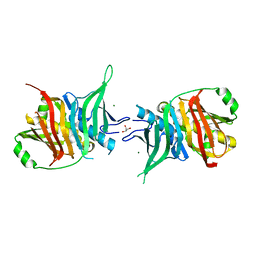 | | Dehydratase domain of module 3 from Brevibacillus Brevis orphan BGC11 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Putative polyketide synthase | | Authors: | Tittes, Y.U, Herbst, D.A, Jakob, R.P, Maier, T. | | Deposit date: | 2022-04-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The structure of a polyketide synthase bimodule core.
Sci Adv, 8, 2022
|
|
7ZSK
 
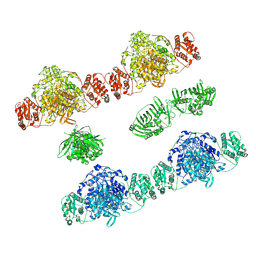 | |
