8UW4
 
 | |
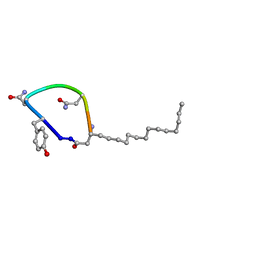
7TZ3
 
 | |
2VBQ
 
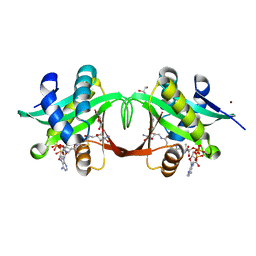 | | Structure of AAC(6')-Iy in complex with bisubstrate analog CoA-S- monomethyl-acetylneamine. | | Descriptor: | (3R,9Z)-17-[(2R,3S,4R,5R,6R)-5-amino-6-{[(1R,2R,3S,4R,6S)-4,6-diamino-2,3-dihydroxycyclohexyl]oxy}-3,4-dihydroxytetrahydro-2H-pyran-2-yl]-3-hydroxy-2,2-dimethyl-4,8,15-trioxo-12-thia-5,9,16-triazaheptadec-9-en-1-yl [(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]methyl dihydrogen diphosphate, AMINOGLYCOSIDE 6'-N-ACETYLTRANSFERASE, GLYCEROL, ... | | Authors: | Vetting, M.W, Magalhaes, M.L, Freiburger, L, Gao, F, Auclair, K, Blanchard, J.S. | | Deposit date: | 2007-09-14 | | Release date: | 2008-01-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Kinetic and Structural Analysis of Bisubstrate Inhibition of the Salmonella Enterica Aminoglycoside 6'-N-Acetyltransferase.
Biochemistry, 47, 2008
|
|
7M77
 
 | |
7M73
 
 | |
7M79
 
 | |
7M67
 
 | |
3RDO
 
 | | Crystal structure of R7-2 streptavidin complexed with biotin | | Descriptor: | BIOTIN, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-01 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
3RE5
 
 | | Crystal structure of R4-6 streptavidin | | Descriptor: | GLYCEROL, PENTAETHYLENE GLYCOL, Streptavidin | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-02 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
3RDS
 
 | | Crystal structure of the refolded R7-2 streptavidin | | Descriptor: | PENTAETHYLENE GLYCOL, Streptavidin | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-01 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
3RDQ
 
 | | Crystal structure of R7-2 streptavidin complexed with desthiobiotin | | Descriptor: | 6-(5-METHYL-2-OXO-IMIDAZOLIDIN-4-YL)-HEXANOIC ACID, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-01 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
3RE6
 
 | | Crystal structure of R4-6 streptavidin | | Descriptor: | GLYCEROL, Streptavidin | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-02 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.823 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
3RDX
 
 | | Crystal structure of ligand-free R7-2 streptavidin | | Descriptor: | GLYCEROL, Streptavidin | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-02 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
3RDM
 
 | | Crystal structure of R7-2 streptavidin complexed with biotin/PEG | | Descriptor: | BIOTIN, PENTAETHYLENE GLYCOL, Streptavidin | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-01 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
3RDU
 
 | | Crystal structure of R7-2 streptavidin complexed with PEG | | Descriptor: | GLYCEROL, PENTAETHYLENE GLYCOL, Streptavidin | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-01 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
