8TPK
 
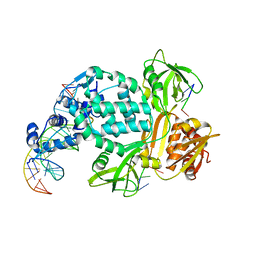 | | P6522 crystal form of C. crescentus DriD-ssDNA-DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*AP*CP*GP*AP*CP*AP*GP*TP*AP*AP*CP*TP*GP*TP*CP*GP*TP*AP*T)-3'), DNA (5'-D(*AP*TP*AP*CP*GP*AP*CP*AP*GP*TP*TP*AP*CP*TP*GP*TP*CP*GP*TP*AP*T)-3'), DNA (5'-D(P*GP*TP*C)-3'), ... | | Authors: | Schumacher, M.A. | | Deposit date: | 2023-08-04 | | Release date: | 2023-11-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structure of the WYL-domain containing transcription activator, DriD, in complex with ssDNA effector and DNA target site.
Nucleic Acids Res., 52, 2024
|
|
6UNX
 
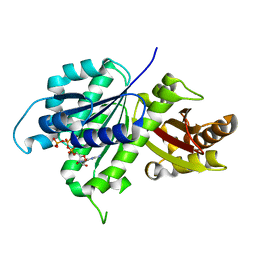 | | Structure of E. coli FtsZ(L178E)-GTP complex | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Schumacher, M.A. | | Deposit date: | 2019-10-13 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution crystal structures of Escherichia coli FtsZ bound to GDP and GTP.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
3M8F
 
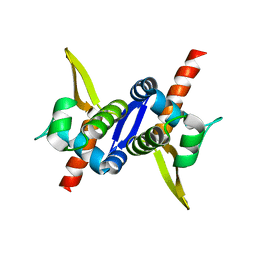 | | Protein structure of type III plasmid segregation TubR mutant | | Descriptor: | Putative DNA-binding protein | | Authors: | Schumacher, M.A, Ni, L. | | Deposit date: | 2010-03-17 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | From the Cover: Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5TZD
 
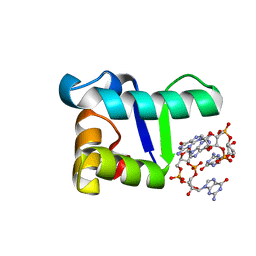 | | Structure of the WT S. venezulae BldD-(CTD-c-di-GMP)2 assembly intermediate | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DNA-binding protein | | Authors: | Schumacher, M. | | Deposit date: | 2016-11-21 | | Release date: | 2017-04-19 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.749 Å) | | Cite: | The Streptomyces master regulator BldD binds c-di-GMP sequentially to create a functional BldD2-(c-di-GMP)4 complex.
Nucleic Acids Res., 45, 2017
|
|
3KZ5
 
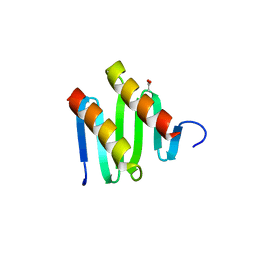 | | Structure of cdomain | | Descriptor: | ACETATE ION, Protein sopB | | Authors: | Schumacher, M.A. | | Deposit date: | 2009-12-07 | | Release date: | 2010-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes.
Nucleic Acids Res., 38, 2010
|
|
3TPT
 
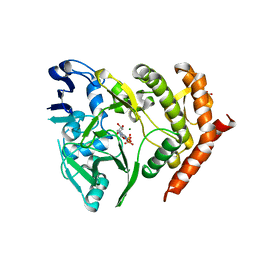 | | Structure of HipA(D309Q) bound to ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | schumacher, M.A, link, T, Brennan, R.G. | | Deposit date: | 2011-09-08 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
5KGO
 
 | | Structure of K. pneumonia MrkH-c-di-GMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Flagellar brake protein YcgR | | Authors: | Schumacher, M. | | Deposit date: | 2016-06-13 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | To be published:
Structures of K. pneumonia MrkH: dual utilization of the PilZ fold for c-di-GMP and DNA binding by a novel activator of biofilm genes
To Be Published
|
|
3M9A
 
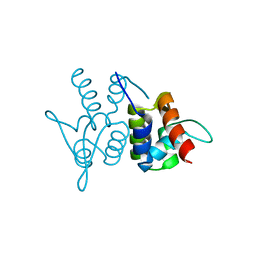 | | Protein structure of type III plasmid segregation TubR | | Descriptor: | Putative DNA-binding protein | | Authors: | Schumacher, M.A, Ni, L. | | Deposit date: | 2010-03-21 | | Release date: | 2010-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | From the Cover: Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3M8K
 
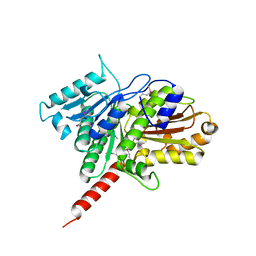 | | Protein structure of type III plasmid segregation TubZ | | Descriptor: | FtsZ/tubulin-related protein | | Authors: | Schumacher, M.A, Ni, L. | | Deposit date: | 2010-03-18 | | Release date: | 2010-07-07 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | From the Cover: Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3DNV
 
 | | MDT Protein | | Descriptor: | DNA (5'-D(*DAP*DCP*DTP*DAP*DTP*DCP*DCP*DCP*DCP*DTP*DTP*DAP*DAP*DGP*DGP*DGP*DGP*DAP*DTP*DAP*DG)-3'), HTH-type transcriptional regulator hipB, Protein hipA, ... | | Authors: | schumacher, M.A. | | Deposit date: | 2008-07-02 | | Release date: | 2009-01-27 | | Last modified: | 2023-04-05 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
3TPB
 
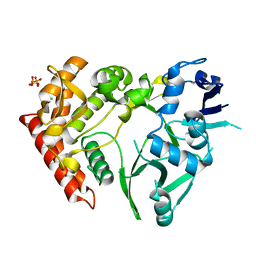 | | Structure of HipA(S150A) | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Serine/threonine-protein kinase HipA | | Authors: | schumacher, M.A. | | Deposit date: | 2011-09-07 | | Release date: | 2012-10-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Role of Unusual P Loop Ejection and Autophosphorylation in HipA-Mediated Persistence and Multidrug Tolerance.
Cell Rep, 2, 2012
|
|
3OQM
 
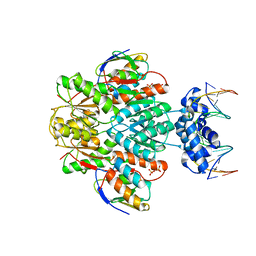 | | structure of ccpa-hpr-ser46p-ackA2 complex | | Descriptor: | 5'-D(*TP*TP*GP*AP*TP*AP*AP*CP*GP*CP*TP*TP*AP*CP*AP*A)-3', 5'-D(*TP*TP*GP*TP*AP*AP*GP*CP*GP*TP*TP*AP*TP*CP*AP*A)-3', Catabolite control protein A, ... | | Authors: | Schumacher, M.A, Sprehe, M, Bartholomae, M, Hillen, W, Brennan, R.G. | | Deposit date: | 2010-09-03 | | Release date: | 2010-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Structures of carbon catabolite protein A-(HPr-Ser46-P) bound to diverse catabolite response element sites reveal the basis for high-affinity binding to degenerate DNA operators.
Nucleic Acids Res., 39, 2011
|
|
1JT0
 
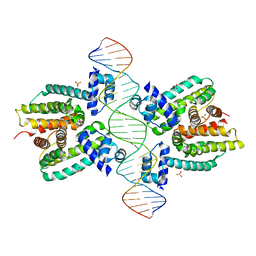 | | Crystal structure of a cooperative QacR-DNA complex | | Descriptor: | HYPOTHETICAL TRANSCRIPTIONAL REGULATOR IN QACA 5'REGION, QACA operator, SULFATE ION | | Authors: | Schumacher, M.A, Miller, M.C, Grkovic, S, Brown, M.H, Skurray, R.A, Brennan, R.G. | | Deposit date: | 2001-08-20 | | Release date: | 2002-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for cooperative DNA binding by two dimers of the multidrug-binding protein QacR.
EMBO J., 21, 2002
|
|
1JTX
 
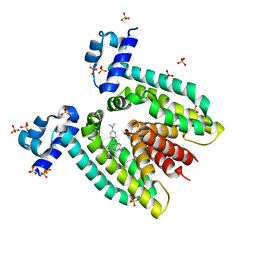 | | Crystal structure of the multidrug binding transcriptional regulator QacR bound to crystal violet | | Descriptor: | CRYSTAL VIOLET, HYPOTHETICAL TRANSCRIPTIONAL REGULATOR IN QACA 5'REGION, SULFATE ION | | Authors: | Schumacher, M.A, Miller, M.C, Grkovic, S, Brown, M.H, Skurray, R.A, Brennan, R.G. | | Deposit date: | 2001-08-22 | | Release date: | 2001-12-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural mechanisms of QacR induction and multidrug recognition.
Science, 294, 2001
|
|
1JUP
 
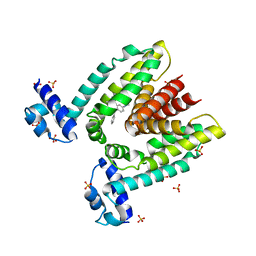 | | Crystal structure of the multidrug binding transcriptional repressor QacR bound to malachite green | | Descriptor: | HYPOTHETICAL TRANSCRIPTIONAL REGULATOR IN QACA 5'REGION, MALACHITE GREEN, SULFATE ION | | Authors: | Schumacher, M.A, Miller, M.C, Grkovic, S, Brown, M.H, Skurray, R.A, Brennan, R.G. | | Deposit date: | 2001-08-24 | | Release date: | 2001-12-12 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural mechanisms of QacR induction and multidrug recognition.
Science, 294, 2001
|
|
2NZU
 
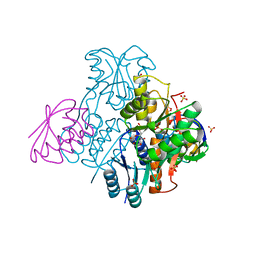 | | Structural mechanism for the fine-tuning of CcpA function by the small molecule effectors G6P and FBP | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, Catabolite control protein, Phosphocarrier protein HPr, ... | | Authors: | Schumacher, M.A, Hillen, W, Brennan, R.G. | | Deposit date: | 2006-11-25 | | Release date: | 2007-05-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Mechanism for the Fine-tuning of CcpA Function by The Small Molecule Effectors Glucose 6-Phosphate and Fructose 1,6-Bisphosphate.
J.Mol.Biol., 368, 2007
|
|
1QX7
 
 | |
1QX5
 
 | |
4E03
 
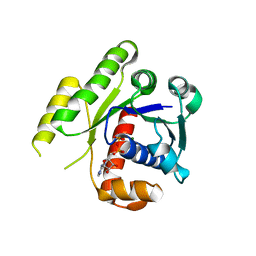 | | Structure of ParF-ADP form 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Plasmid partitioning protein ParF | | Authors: | Schumacher, M.A, Ye, Q, Barge, M.R, Barilla, D, Hayes, F. | | Deposit date: | 2012-03-02 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Mechanism of ATP-induced Polymerization of the Partition Factor ParF: IMPLICATIONS FOR DNA SEGREGATION.
J.Biol.Chem., 287, 2012
|
|
1DL7
 
 | |
3PM1
 
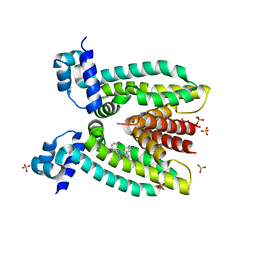 | | Structure of QacR E90Q bound to Ethidium | | Descriptor: | ETHIDIUM, HTH-type transcriptional regulator qacR, SULFATE ION | | Authors: | Schumacher, M.A. | | Deposit date: | 2010-11-15 | | Release date: | 2011-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A single acidic residue can guide binding site selection but does not govern QacR cationic-drug affinity.
Plos One, 6, 2011
|
|
2PUC
 
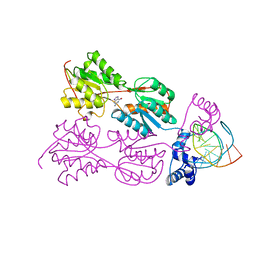 | | CRYSTAL STRUCTURE OF THE LACI FAMILY MEMBER, PURR, BOUND TO DNA: MINOR GROOVE BINDING BY ALPHA HELICES | | Descriptor: | DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*CP*GP*T )-3'), GUANINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Schumacher, M.A, Choi, K.Y, Zalkin, H, Brennan, R.G. | | Deposit date: | 1997-10-04 | | Release date: | 1998-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of LacI member, PurR, bound to DNA: minor groove binding by alpha helices.
Science, 266, 1994
|
|
2PUD
 
 | | CRYSTAL STRUCTURE OF THE LACI FAMILY MEMBER, PURR, BOUND TO DNA: MINOR GROOVE BINDING BY ALPHA HELICES | | Descriptor: | DNA (5'-D(*TP*AP*CP*GP*CP*AP*AP*AP*CP*GP*TP*TP*TP*GP*CP*GP*T )-3'), HYPOXANTHINE, PROTEIN (PURINE REPRESSOR) | | Authors: | Schumacher, M.A, Choi, K.Y, Zalkin, H, Brennan, R.G. | | Deposit date: | 1997-10-04 | | Release date: | 1998-05-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of LacI member, PurR, bound to DNA: minor groove binding by alpha helices.
Science, 266, 1994
|
|
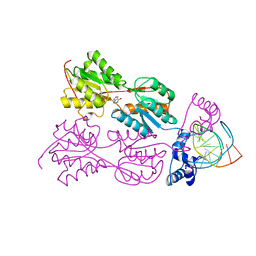
3HZI
 
 | | Structure of mdt protein | | Descriptor: | 5'-D(*DAP*DCP*DTP*DAP*DTP*DCP*DCP*DCP*DCP*DTP*DTP*DAP*DAP*DGP*DGP*DGP*DGP*DAP*DTP*DAP*DG)-3', ADENOSINE-5'-TRIPHOSPHATE, HTH-type transcriptional regulator hipB, ... | | Authors: | Schumacher, M.A. | | Deposit date: | 2009-06-23 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB.
Science, 323, 2009
|
|
4DZZ
 
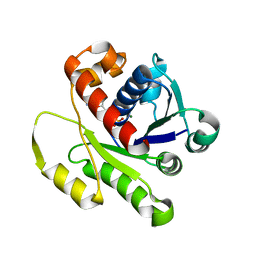 | | Structure of ParF-ADP, crystal form 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Plasmid partitioning protein ParF | | Authors: | Schumacher, M.A, Ye, Q, Barge, M.R, Barilla, D, Hayes, F. | | Deposit date: | 2012-03-01 | | Release date: | 2012-06-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Mechanism of ATP-induced Polymerization of the Partition Factor ParF: IMPLICATIONS FOR DNA SEGREGATION.
J.Biol.Chem., 287, 2012
|
|
