8CRF
 
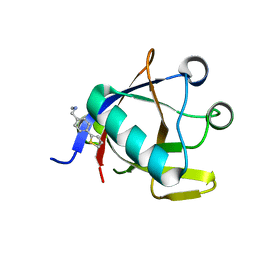 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 5E11 refined against anomalous diffraction data | | Descriptor: | Host translation inhibitor nsp1, ~{N}-methyl-1-(4-thiophen-2-ylphenyl)methanamine | | Authors: | Ma, S, Mykhaylyk, V, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2023-03-08 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | High-Confidence Placement of Fragments into Electron Density Using Anomalous Diffraction-A Case Study Using Hits Targeting SARS-CoV-2 Non-Structural Protein 1.
Int J Mol Sci, 24, 2023
|
|
8CRM
 
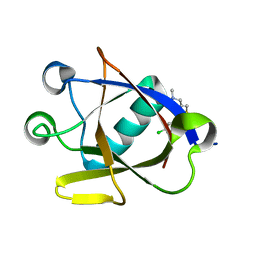 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 11C6 refined against anomalous diffraction data | | Descriptor: | 1-[2-(3-chlorophenyl)-1,3-thiazol-4-yl]-~{N}-methyl-methanamine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Mikhailik, V, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2023-03-08 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | High-Confidence Placement of Fragments into Electron Density Using Anomalous Diffraction-A Case Study Using Hits Targeting SARS-CoV-2 Non-Structural Protein 1.
Int J Mol Sci, 24, 2023
|
|
8CRK
 
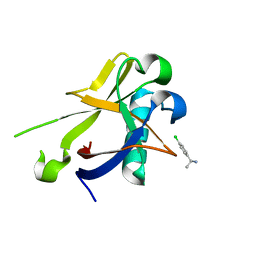 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 7H2 refined against anomalous diffraction data | | Descriptor: | (1~{R})-1-(4-chlorophenyl)ethanamine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Mikhailik, V, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2023-03-08 | | Release date: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-Confidence Placement of Fragments into Electron Density Using Anomalous Diffraction-A Case Study Using Hits Targeting SARS-CoV-2 Non-Structural Protein 1.
Int J Mol Sci, 24, 2023
|
|
7DFV
 
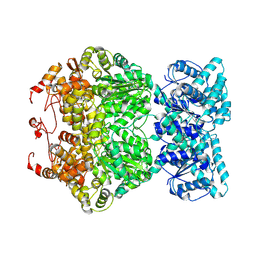 | | Cryo-EM structure of plant NLR RPP1 tetramer core part | | Descriptor: | NAD+ hydrolase (NADase) | | Authors: | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | Deposit date: | 2020-11-10 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
6PB0
 
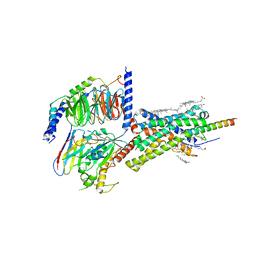 | | Cryo-EM structure of Urocortin 1-bound Corticotropin-releasing factor 1 receptor in complex with Gs protein and Nb35 | | Descriptor: | CHOLESTEROL, Corticotropin-releasing factor receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ma, S, Shen, Q, Zhao, L.-H, Mao, C, Zhou, X.E, Shen, D.-D, de Waal, P.W, Bi, P, Li, C, Jiang, Y, Wang, M.-W, Sexton, P.M, Wootten, D, Melcher, K, Zhang, Y, Xu, H.E. | | Deposit date: | 2019-06-12 | | Release date: | 2020-02-12 | | Last modified: | 2020-02-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular Basis for Hormone Recognition and Activation of Corticotropin-Releasing Factor Receptors.
Mol.Cell, 77, 2020
|
|
6PB1
 
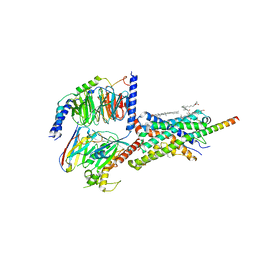 | | Cryo-EM structure of Urocortin 1-bound Corticotropin-releasing factor 2 receptor in complex with Gs protein and Nb35 | | Descriptor: | CHOLESTEROL, Corticotropin-releasing factor receptor 2, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ma, S, Shen, Q, Zhao, L.-H, Mao, C, Zhou, X.E, Shen, D.-D, de Waal, P.W, Bi, P, Li, C, Jiang, Y, Wang, M.-W, Sexton, P.M, Wootten, D, Melcher, K, Zhang, Y, Xu, H.E. | | Deposit date: | 2019-06-12 | | Release date: | 2020-02-12 | | Last modified: | 2020-02-19 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular Basis for Hormone Recognition and Activation of Corticotropin-Releasing Factor Receptors.
Mol.Cell, 77, 2020
|
|
8AYS
 
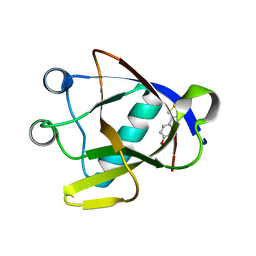 | | SARS-CoV-2 non-structural protein-1 (nsp1) in complex with 4-(2-aminothiazol-4-yl)phenol | | Descriptor: | 4-(2-amino-1,3-thiazol-4-yl)phenol, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2022-09-03 | | Release date: | 2022-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Two Ligand-Binding Sites on SARS-CoV-2 Non-Structural Protein 1 Revealed by Fragment-Based X-ray Screening.
Int J Mol Sci, 23, 2022
|
|
8RF2
 
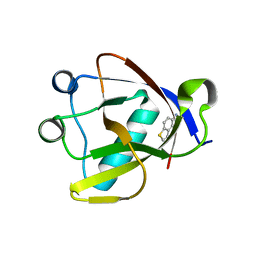 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 1E7 refined against the anomalous diffraction data | | Descriptor: | 1-benzothiophen-5-amine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RF6
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 11A7_AL5 refined against the anomalous diffraction data | | Descriptor: | 6-iodanyl-2,3-dihydro-1,3-benzothiazol-2-amine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RFF
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 6A6 refined against the anomalous diffraction data | | Descriptor: | 1,3-benzothiazol-2-amine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RF8
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 11A7_AL6 refined against the anomalous diffraction data | | Descriptor: | 6-bromanyl-1,3-benzothiazol-2-amine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RF3
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 7G3 refined against the anomalous diffraction data | | Descriptor: | 2-(1-benzothiophen-3-yl)ethanoic acid, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RF5
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 11A7 refined against the anomalous diffraction data | | Descriptor: | 6-fluoro-1,3-benzothiazol-2-amine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8AZ8
 
 | | SARS-CoV-2 non-structural protein-1 (nsp1) in complex with 2-(benzylamino)ethan-1-ol | | Descriptor: | 2-[(phenylmethyl)amino]ethanol, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Pinotsis, N, Bowler, M.W, Kozielski, F. | | Deposit date: | 2022-09-05 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Two Ligand-Binding Sites on SARS-CoV-2 Non-Structural Protein 1 Revealed by Fragment-Based X-ray Screening.
Int J Mol Sci, 23, 2022
|
|
8RF4
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 9D4 refined against the anomalous diffraction data | | Descriptor: | 4-chloranyl-1~{H}-indazol-3-amine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RFD
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 7H2_AL2 refined against the anomalous diffraction data | | Descriptor: | (1~{R})-1-(4-iodophenyl)ethanamine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RFC
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 7H2_AL1 refined against the anomalous diffraction data | | Descriptor: | (1~{R})-1-(4-bromophenyl)ethanamine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8A55
 
 | |
7F4H
 
 | | Cryo-EM structure of afamelanotide-bound melanocortin-1 receptor in complex with Gs protein, Nb35 and scFv16 | | Descriptor: | Afamelanotide, CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ma, S, Chen, Y, Dai, A, Yin, W, Guo, J, Yang, D, Zhou, F, Jiang, Y, Wang, M.-W, Xu, H.E. | | Deposit date: | 2021-06-18 | | Release date: | 2021-09-08 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mechanism of calcium-mediated hormone recognition and G beta interaction by the human melanocortin-1 receptor.
Cell Res., 31, 2021
|
|
7F4I
 
 | | Cryo-EM structure of SHU9119-bound melanocortin-1 receptor in complex with Gs protein and Nb35 | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ma, S, Chen, Y, Dai, A, Yin, W, Guo, J, Yang, D, Zhou, F, Jiang, Y, Wang, M.-W, Xu, H.E. | | Deposit date: | 2021-06-18 | | Release date: | 2021-09-08 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural mechanism of calcium-mediated hormone recognition and G beta interaction by the human melanocortin-1 receptor.
Cell Res., 31, 2021
|
|
7F4D
 
 | | Cryo-EM structure of alpha-MSH-bound melanocortin-1 receptor in complex with Gs protein and Nb35 | | Descriptor: | CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ma, S, Chen, Y, Dai, A, Yin, W, Guo, J, Yang, D, Zhou, F, Jiang, Y, Wang, M.-W, Xu, H.E. | | Deposit date: | 2021-06-18 | | Release date: | 2021-09-08 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural mechanism of calcium-mediated hormone recognition and G beta interaction by the human melanocortin-1 receptor.
Cell Res., 31, 2021
|
|
7F4F
 
 | | Cryo-EM structure of afamelanotide-bound melanocortin-1 receptor in complex with Gs protein and scFv16 | | Descriptor: | Afamelanotide, CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Ma, S, Chen, Y, Dai, A, Yin, W, Guo, J, Yang, D, Zhou, F, Jiang, Y, Wang, M.-W, Xu, H.E. | | Deposit date: | 2021-06-18 | | Release date: | 2021-09-08 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mechanism of calcium-mediated hormone recognition and G beta interaction by the human melanocortin-1 receptor.
Cell Res., 31, 2021
|
|
7CRC
 
 | | Cryo-EM structure of plant NLR RPP1 tetramer in complex with ATR1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Avirulence protein ATR1, ... | | Authors: | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | Deposit date: | 2020-08-13 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
7CRB
 
 | | Cryo-EM structure of plant NLR RPP1 LRR-ID domain in complex with ATR1 | | Descriptor: | Avirulence protein ATR1, NAD+ hydrolase (NADase) | | Authors: | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | Deposit date: | 2020-08-13 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
2KW1
 
 | | Solution structure of CTD | | Descriptor: | Fas apoptotic inhibitory molecule 1 | | Authors: | Ma, S, Zheng, X, Jin, C. | | Deposit date: | 2010-03-30 | | Release date: | 2011-04-13 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of CTD
To be Published
|
|
