3T4H
 
 | | Crystal Structure of AlkB in complex with Fe(III) and N-Oxalyl-S-(3-nitrobenzyl)-L-cysteine | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Alpha-ketoglutarate-dependent dioxygenase AlkB, FE (III) ION, ... | | Authors: | Ma, J, Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2011-07-26 | | Release date: | 2012-03-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Dynamic combinatorial mass spectrometry leads to inhibitors of a 2-oxoglutarate-dependent nucleic Acid demethylase.
J.Med.Chem., 55, 2012
|
|
6UZ1
 
 | |
7EZX
 
 | |
7C7U
 
 | | Biofilm associated protein - BSP domain | | Descriptor: | Biofilm-associated surface protein, CALCIUM ION | | Authors: | Ma, J.F, Xu, Z.H, Zhang, Y.K, Cheng, X, Fan, S.L, Wang, J.W, Fang, X.Y. | | Deposit date: | 2020-05-26 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural mechanism for modulation of functional amyloid and biofilm formation by Staphylococcal Bap protein switch.
Embo J., 40, 2021
|
|
7C7R
 
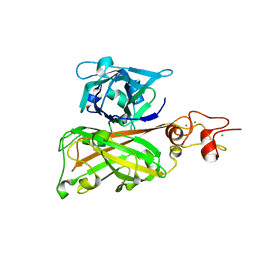 | | Biofilm associated protein - B domain | | Descriptor: | Biofilm-associated surface protein, CALCIUM ION | | Authors: | Ma, J.F, Xu, Z.H, Zhang, Y.K, Cheng, X, Fan, S.L, Wang, J.W, Fang, X.Y. | | Deposit date: | 2020-05-26 | | Release date: | 2021-05-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.07 Å) | | Cite: | Structural mechanism for modulation of functional amyloid and biofilm formation by Staphylococcal Bap protein switch.
Embo J., 40, 2021
|
|
1O5W
 
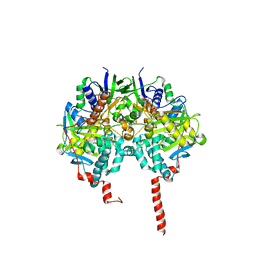 | | The structure basis of specific recognitions for substrates and inhibitors of rat monoamine oxidase A | | Descriptor: | Amine oxidase [flavin-containing] A, FLAVIN-ADENINE DINUCLEOTIDE, N-[3-(2,4-DICHLOROPHENOXY)PROPYL]-N-METHYL-N-PROP-2-YNYLAMINE | | Authors: | Ma, J, Yoshimura, M, Yamashita, E, Nakagawa, A, Ito, A, Tsukihara, T. | | Deposit date: | 2003-10-06 | | Release date: | 2004-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure of rat monoamine oxidase a and its specific recognitions for substrates and inhibitors.
J.Mol.Biol., 338, 2004
|
|
2QDW
 
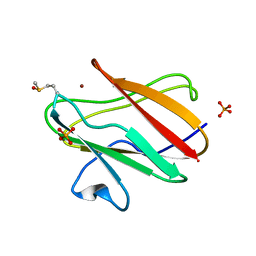 | | Structure of Cu(I) form of the M51A mutant of amicyanin | | Descriptor: | Amicyanin, COPPER (I) ION, PHOSPHATE ION | | Authors: | Ma, J.K, Wang, Y, Carrell, C.J, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2007-06-21 | | Release date: | 2007-12-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | A single methionine residue dictates the kinetic mechanism of interprotein electron transfer from methylamine dehydrogenase to amicyanin.
Biochemistry, 46, 2007
|
|
1YTU
 
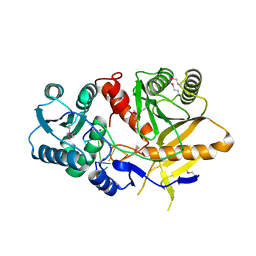 | | Structural basis for 5'-end-specific recognition of the guide RNA strand by the A. fulgidus PIWI protein | | Descriptor: | 5'-R(P*AP*GP*AP*CP*AP*G)-3', 5'-R(P*UP*GP*UP*C)-3', MAGNESIUM ION, ... | | Authors: | Ma, J.B, Yuan, Y.R, Meister, G, Pei, Y, Tuschl, T, Patel, D.J. | | Deposit date: | 2005-02-11 | | Release date: | 2005-04-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for 5'-end-specific recognition of guide RNA by the A. fulgidus Piwi protein.
Nature, 434, 2005
|
|
3U4S
 
 | | Histone Lysine demethylase JMJD2A in complex with T11C peptide substrate crosslinked to N-oxalyl-D-cysteine | | Descriptor: | HISTONE 3 TAIL ANALOG (T11C Peptide), Lysine-specific demethylase 4A, N-(carboxycarbonyl)-D-cysteine, ... | | Authors: | Ma, J, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2011-10-10 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Linking of 2-Oxoglutarate and Substrate Binding Sites Enables Potent and Highly Selective Inhibition of JmjC Histone Demethylases.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
6CSF
 
 | | Crystal structure of sodium/alanine symporter AgcS with D-alanine bound | | Descriptor: | D-ALANINE, Monoclonal antibody FAB heavy chain, Monoclonal antibody FAB light chain, ... | | Authors: | Ma, J, Reyes, F.E, Gonen, T. | | Deposit date: | 2018-03-20 | | Release date: | 2019-01-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for substrate binding and specificity of a sodium-alanine symporter AgcS.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
6CSE
 
 | | Crystal structure of sodium/alanine symporter AgcS with L-alanine bound | | Descriptor: | ALANINE, Monoclonal antibody FAB heavy chain, Monoclonal antibody FAB light chain, ... | | Authors: | Ma, J, Reyes, F.E, Gonen, T. | | Deposit date: | 2018-03-20 | | Release date: | 2019-01-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Structural basis for substrate binding and specificity of a sodium-alanine symporter AgcS.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
8ZSZ
 
 | |
8ZSB
 
 | |
3CZ3
 
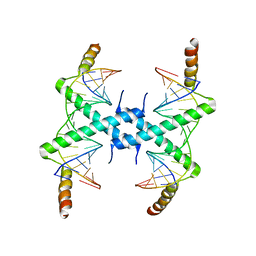 | | Crystal structure of Tomato Aspermy Virus 2b in complex with siRNA | | Descriptor: | Protein 2b, RNA (5'-R(P*CP*GP*UP*AP*CP*GP*CP*GP*GP*AP*AP*UP*AP*CP*UP*UP*CP*GP*A)-3'), RNA (5'-R(P*UP*CP*GP*AP*AP*GP*UP*AP*UP*UP*CP*CP*GP*CP*GP*UP*AP*CP*G)-3') | | Authors: | Ma, J.B, Li, F, Ding, S.W, Patel, D.J. | | Deposit date: | 2008-04-27 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.23 Å) | | Cite: | Structural Basis for siRNA Recognition by 2b, a Viral Suppressor of Non-Cell Autonomous RNA Silencing
To be Published
|
|
8J4N
 
 | |
8J4O
 
 | |
2LR1
 
 | |
9ASF
 
 | |
9ASG
 
 | |
7RRG
 
 | |
7EKA
 
 | | crystal structure of epigallocatechin binding with alpha-lactalbumin | | Descriptor: | 2-(3,4,5-TRIHYDROXY-PHENYL)-CHROMAN-3,5,7-TRIOL, Alpha-lactalbumin | | Authors: | Ma, J, Yao, Q, Chen, X, Zang, J. | | Deposit date: | 2021-04-05 | | Release date: | 2023-11-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Weak Binding of Epigallocatechin to alpha-Lactalbumin Greatly Improves Its Stability and Uptake by Caco-2 Cells.
J.Agric.Food Chem., 69, 2021
|
|
2GB2
 
 | | The P52G mutant of amicyanin in the Cu(II) state. | | Descriptor: | Amicyanin, COPPER (II) ION | | Authors: | Ma, J.K, Carrell, C.J, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2006-03-09 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Site-Directed Mutagenesis of Proline 52 To Glycine in Amicyanin Converts a True Electron Transfer Reaction into One that Is Conformationally Gated.
Biochemistry, 45, 2006
|
|
3MIL
 
 | | Crystal structure of isoamyl acetate-hydrolyzing esterase from Saccharomyces cerevisiae | | Descriptor: | GLYCEROL, Isoamyl acetate-hydrolyzing esterase | | Authors: | Ma, J, Lu, Q, Yuan, Y, Li, K, Ge, H, Go, Y, Niu, L, Teng, M. | | Deposit date: | 2010-04-11 | | Release date: | 2010-11-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of isoamyl acetate-hydrolyzing esterase from Saccharomyces cerevisiae reveals a novel active site architecture and the basis of substrate specificity
Proteins, 79, 2011
|
|
2GBA
 
 | | Reduced Cu(I) form at pH 4 of P52G mutant of amicyanin | | Descriptor: | COPPER (I) ION, amicyanin | | Authors: | Ma, J.K, Carrell, C.J, Mathews, F.S, Davidson, V.L. | | Deposit date: | 2006-03-10 | | Release date: | 2006-08-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (0.92 Å) | | Cite: | Site-Directed Mutagenesis of Proline 52 To Glycine in Amicyanin Converts a True Electron Transfer Reaction into One that Is Conformationally Gated.
Biochemistry, 45, 2006
|
|
6AKM
 
 | | Crystal structure of SLMAP-SIKE1 complex | | Descriptor: | GLYCEROL, Sarcolemmal membrane-associated protein, Suppressor of IKBKE 1 | | Authors: | Ma, J, Chen, M, Zhou, Z.C. | | Deposit date: | 2018-09-02 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Architecture, substructures, and dynamic assembly of STRIPAK complexes in Hippo signaling.
Cell Discov, 5, 2019
|
|
